PROBLEMS
PROBLEMS
Question 5.1
It’s not the heat … Why is Taq polymerase especially useful for PCR?
Question 5.2
The right template. Ovalbumin is the major protein of egg white. The chicken ovalbumin gene contains eight exons separated by seven introns. Should ovalbumin cDNA or ovalbumin genomic DNA be used to form the protein in E. coli? Why?
Question 5.3
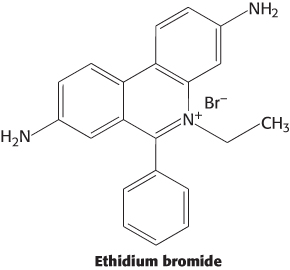
Handle with care. Ethidium bromide is a commonly used stain for DNA molecules after separation by gel electrophoresis. The chemical structure of ethidium bromide is shown here. Based on this structure, suggest how this stain binds to DNA.
Question 5.4
Cleavage frequency. The restriction enzyme AluI cleaves at the sequence 5′-AGCT-
Question 5.5
The right cuts. Suppose that a human genomic library is prepared by exhaustive digestion of human DNA with the EcoRI restriction enzyme. Fragments averaging about 4 kb in length would be generated. Is this procedure suitable for cloning large genes? Why or why not?
Question 5.6
A revealing cleavage. Sickle-
Question 5.7
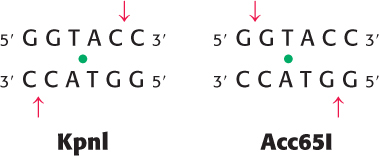
Sticky ends? The restriction enzymes KpnI and Acc65I recognize and cleave the same 6-
167
Question 5.8
Many melodies from one cassette. Suppose that you have isolated an enzyme that digests paper pulp and have obtained its cDNA. The goal is to produce a mutant that is effective at high temperature. You have engineered a pair of unique restriction sites in the cDNA that flank a 30-
Question 5.9
A blessing and a curse. The power of PCR can also create problems. Suppose someone claims to have isolated dinosaur DNA by using PCR. What questions might you ask to determine if it is indeed dinosaur DNA?
Question 5.10
Rich or poor? DNA sequences that are highly enriched in G–
Question 5.11
Questions of accuracy. The stringency of PCR amplification can be controlled by altering the temperature at which the primers and the target DNA undergo hybridization. How would altering the temperature of hybridization affect the amplification? Suppose that you have a particular yeast gene A and that you wish to see if it has a counterpart in humans. How would controlling the stringency of the hybridization help you?
Question 5.12
Terra incognita. PCR is typically used to amplify DNA that lies between two known sequences. Suppose that you want to explore DNA on both sides of a single known sequence. Devise a variation of the usual PCR protocol that would enable you to amplify entirely new genomic terrain.
Question 5.13
A puzzling ladder. A gel pattern displaying PCR products shows four strong bands. The four pieces of DNA have lengths that are approximately in the ratio of 1 : 2 : 3 : 4. The largest band is cut out of the gel, and PCR is repeated with the same primers. Again, a ladder of four bands is evident in the gel. What does this result reveal about the structure of the encoded protein?
Question 5.14
Chromosome walking. Propose a method for isolating a DNA fragment that is adjacent in the genome to a previously isolated DNA fragment. Assume that you have access to a complete library of DNA fragments in a BAC vector but that the sequence of the genome under study has not yet been determined.
Question 5.15
Probe design. Which of the following amino acid sequences would yield the most optimal oligonucleotide probe?

Question 5.16
Man’s best friend. Why might the genomic analysis of dogs be particularly useful for investigating the genes responsible for body size and other physical characteristics?
Question 5.17
Of mice and men. You have identified a gene that is located on human chromosome 20 and wish to identify its location within the mouse genome. On which chromosome would you be most likely to find the mouse counterpart of this gene?
Chapter Integration Problems
Question 5.18
Designing primers I. A successful PCR experiment often depends on designing the correct primers. In particular, the Tm for each primer should be approximately the same. What is the basis of this requirement?
Question 5.19
Designing primers II. You wish to amplify a segment of DNA from a plasmid template by PCR with the use of the following primers: 5′-GGATCGATGCTCGCGA-
Chapter Integration and Data Interpretation Problem
Question 5.20
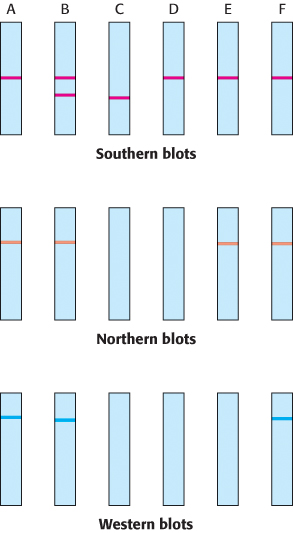
Any direction but east. A series of people are found to have difficulty eliminating certain types of drugs from their bloodstreams. The problem has been linked to a gene X, which encodes an enzyme Y. Six people were tested with the use of various techniques of molecular biology. Person A is a normal control, person B is asymptomatic but some of his children have the metabolic problem, and persons C through F display the trait. Tissue samples from each person were obtained. Southern analysis was performed on the DNA after digestion with the restriction enzyme HindIII. Northern analysis of mRNA also was done. In both types of analysis, the gels were probed with labeled X cDNA. Finally, a western blot with an enzyme-
168
Data Interpretation Problems
Question 5.21
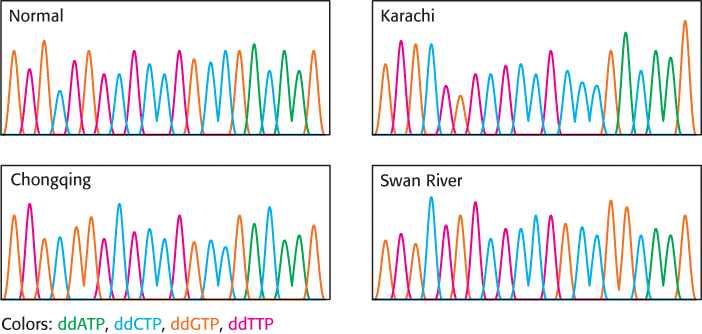
DNA diagnostics. Representations of sequencing chromatograms for variants of the α chain of human hemoglobin are shown here. What is the nature of the amino acid change in each of the variants? The first triplet encodes valine.
Question 5.22
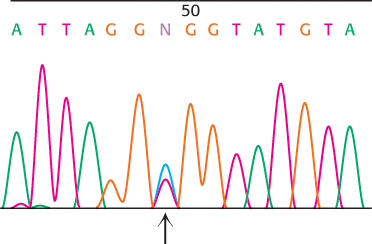
Two peaks. In the course of studying a gene and its possible mutation in humans, you obtain genomic DNA samples from a collection of persons and PCR amplify a region of interest within this gene. For one of the samples, you obtain the sequencing chromatogram shown here. Provide an explanation for the appearance of these data at position 49 (indicated by the arrow):
 Visit www.whfreeman.com/
Visit www.whfreeman.com/