Chapter 5
Chapter 5
1. Taq polymerase is the DNA polymerase from the thermophilic bacterium that lives in hot springs. Consequently, it is heat stable and can withstand the high temperatures required for PCR without denaturing.
2. Ovalbumin cDNA should be used. E. coli lacks the machinery to splice the primary transcript arising from genomic DNA.
3. Consistent with its planar, aromatic structure, ethidium bromide is a DNA intercalator: it aligns itself between the paired bases in a DNA duplex.
4. The presence of the AluI sequence would, on average, be (1/4)4, or 1/256, because the likelihood of any base being at any position is one-
5. No, because most human genes are much longer than 4 kb. A fragment would contain only a small part of a complete gene.
6. Southern blotting of an MstII digest would distinguish between the normal and the mutant genes. The loss of a restriction site would lead to the replacement of two fragments on the Southern blot by a single longer fragment. Such a finding would not prove that GTG replaced GAG; other sequence changes at the restriction site could yield the same result.
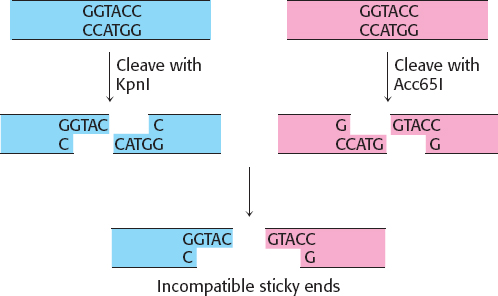
7. Although the two enzymes cleave the same recognition site, they each break different bonds within the 6-
8. A simple strategy for generating many mutants is to synthesize a degenerate set of cassettes by using a mixture of activated nucleosides in particular rounds of oligonucleotide synthesis. Suppose that the 30-
9. Because PCR can amplify as little as one molecule of DNA, statements claiming the isolation of ancient DNA need to be greeted with some skepticism. The DNA would need to be sequenced. Is it similar to human, bacterial, or fungal DNA? If so, contamination is the likely source of the amplified DNA. Is it similar to that of birds or crocodiles? This sequence similarity would strengthen the case that it is dinosaur DNA because these species are evolutionarily close to dinosaurs.
10. PCR amplification is greatly hindered by the presence of G–
11. At high temperatures of hybridization, only very close matches between primer and target would be stable because all (or most) of the bases would need to find partners to stabilize the primer–
12. Digest genomic DNA with a restriction enzyme, and select the fragment that contains the known sequence. Circularize this fragment. Then carry out PCR with the use of a pair of primers that serve as templates for the synthesis of DNA away from the known sequence.
13. The encoded protein contains four repeats of a specific sequence.
14. Use chemical synthesis or the polymerase chain reaction to prepare hybridization probes that are complementary to both ends of the known (previously isolated) DNA fragment. Challenge clones representing the library of DNA fragments with both of the hybridization probes. Select clones that hybridize to one of the probes but not the other; such clones are likely to represent DNA fragments that contain one end of the known fragment along with the adjacent region of the particular chromosome.
15. The codon(s) for each amino acid can be used to determine the number of possible nucleotide sequences that encode each peptide sequence (Table 4.5):
Ala–
4 × 1 × 6 × 6 × 4 × 1 = 576 total sequences
Gly–
4 × 1 × 2 × 1 × 2 × 2 = 32 total sequences
Cys–
2 × 4 × 1 × 2 × 2 × 3 = 96 total sequences
Arg–
6 × 6 × 1 × 6 × 2 × 2 = 864 total sequences
The set of DNA sequences encoding the peptide Gly-
16. Within a single species, individual dogs show enormous variation in body size and substantial diversity in other physical characteristics. Therefore, genomic analysis of individual dogs would provide valuable clues concerning the genes responsible for the diversity within the species.
17. On the basis of the comparative genome map shown in Figure 5.28, the region of greatest overlap with human chromosome 20 can be found on mouse chromosome 2.
18. Tm is the melting temperature of a double-
A7
19. Careful comparison of the sequences reveals that there is a 7-
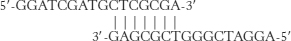
In a PCR experiment, these primers would likely anneal to one another, preventing their interaction with the template DNA. During DNA synthesis by the polymerase, each primer would act as a template for the other primer, leading to the amplification of a 25-
20. A mutation in person B has altered one of the alleles for gene X, leaving the other intact. The fact that the mutated allele is smaller suggests that a deletion has occurred in one copy of the gene. The one functioning copy is transcribed and translated and apparently produces enough protein to render the person asymptomatic.
Person C has only the smaller version of the gene. This gene is neither transcribed (negative northern blot) nor translated (negative western blot).
Person D has a normal-
Person E has a normal-
Person F has a normal amount of protein but still displays the metabolic problem. This finding suggests that the mutation affects the activity of the protein—
21. Chongqing: residue 2, L → R, CTG → CGG
Karachi: residue 5, A → P, GCC → CCC
Swan River: residue 6, D → G, GAC → GGC
22. This particular person is heterozygous for this particular mutation: one allele is wild type, whereas the other carries a point mutation at this position. Both alleles are PCR amplified in this experiment, yielding the “dual peak” appearance on the sequencing chromatogram.