Chapter 8
Chapter 8
1. Rate enhancement and substrate specificity.
2. A cofactor.
3. Coenzymes and metals.
A9
4. Vitamins are converted into coenzymes.
5. Enzymes facilitate the formation of the transition state.
6. The intricate three-
7. The energy required to reach the transition state (the activation energy) is returned when the transition state proceeds to product.
8. (a) 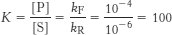 . Using equation 5 in the text, ΔG°′ = − 11.42 kJ mol−1 (−2.73 kcal mol−1)
. Using equation 5 in the text, ΔG°′ = − 11.42 kJ mol−1 (−2.73 kcal mol−1)
(b) kF = 10−2 s−1 and kR = 10−4 s−1. The equilibrium constant and ΔG°′ values are the same for both the uncatalyzed and catalyzed reactions.
9. Protein hydrolysis has a large activation energy. Protein synthesis must require energy to proceed.
10. The enzymes help protect the fluid that surrounds eyes from bacterial infection.
11. Binding energy is the free energy released when two molecules bind together, such as when an enzyme and a substrate interact.
12. Binding energy is maximized when an enzyme interacts with the transition state, thereby facilitating the formation of the transition state and enhancing the rate of the reaction.
13. There would be no catalytic activity. If the enzyme–
14. Transition states are very unstable. Consequently, molecules that resemble transition states are themselves likely to be unstable and, hence, difficult to synthesize.
15. (a) 0; (b) +28.53; (c) −22.84; (d) −11.42; (e) +5.69.
16. (a) 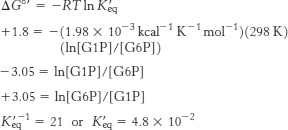
Because [G6P]/[G1P] = 21, there is 1 molecule of G1P for every 21 molecules of G6P. Because we started with 0.1 M, the [G1P] is 1/22(0.1 M) = 0.0045 M and [G6P] must be 21/22(0.1 M) or 0.096 M. Consequently, the reaction does not proceed as written to a significant extent.
(b) Supply G6P at a high rate and remove G1P at a high rate by other reactions. In other words, make sure that the [G6P]/[G1P] is kept large.
17. Keq = 19, ΔG°′ = −7.3 kJ mol−1 (−1.77 kcal mol−1)
18. The three-
19. At substrate concentrations near the KM, the enzyme displays significant catalysis yet is sensitive to changes in substrate concentration.
20. No, KM is not equal to the dissociation constant, because the numerator also contains k2, the rate constant for the conversion of the enzyme–
21. When [S] = 10 KM, V0 = 0.91 Vmax. When [S] = 20 KM, V0 = 95 Vmax.
So any Michaelis–
22. (a) 31.1 μmol; (b) 0.05 μmol; (c) 622 s−1, a midrange value for enzymes (Table 8.5).
23. (a) Yes, KM = 5.2 × 10−6 M; (b) Vmax = 6.8 × 10−10 mol minute−1; (c) 337 s−1.
24. Penicillinase, like glycopeptide transpeptidase, forms an acyl-
25. (a) Vmax is 9.5 μmol minute−1. KM is 1.1 × 10−5 M, the same as without inhibitor.
(b) Noncompetitive.
(c) 2.5 × 10−5 M
(d) fES = 0.73, in the presence or absence of this noncompetitive inhibitor.
26. (a) V0 = Vmax − (V0/[S]) KM
(b) Slope = −KM, y-intercept = Vmax, x-intercept = Vmax/KM
(c) An Eadie–
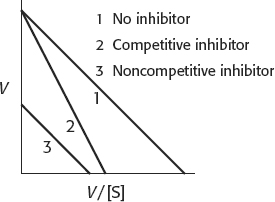
27. Sequential reactions are characterized by the formation of a ternary complex consisting of the enzyme and both substrates. Double-
28. The rates of utilization of substrates A and B are given by
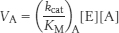
and
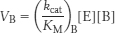
Hence, the ratio of these rates is
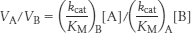
Thus, an enzyme discriminates between competing substrates on the basis of their values of kcat/KM rather than of KM alone.
29. The mutation slows the reaction by a factor of 100 because the activation free energy is increased by +11.42 kJ mol−1 (+2.73 kcal mol−1). Strong binding of the substrate relative to the transition state slows catalysis.
30. 11 μmol minute−1
31. (a) This piece of information is necessary for determining the correct dosage of succinylcholine to administer.
(b) The duration of the paralysis depends on the ability of the serum cholinesterase to clear the drug. If there were one-
(c) KM is the concentration needed by the enzyme to reach ½ Vmax. Consequently, for a given concentration of substrate, the reaction catalyzed by the enzyme with the lower KM will have the higher rate. The mutant patient with the higher KM will clear the drug at a much lower rate.
32. (a) KM is a measure of affinity only if k2 is rate limiting, which is the case here. Therefore, the lower KM means higher affinity. The mutant enzyme has a higher affinity.
A10
(b) 50 μmol minute−1. 10 mM is KM, and KM yields ½ Vmax. Vmax is 100 μmol minute−1, and so …
(c) Enzymes do not alter the equilibrium of the reaction.
33. Enzyme 2. Despite the fact that enzyme 1 has a higher Vmax than enzyme 2, enzyme 2 shows greater activity at the concentration of the substrate in the environment because enzyme 2 has a lower KM for the substrate.
34. (a) The most effective means of measuring the efficiency of any enzyme–
(b) The kcat/KM for this substrate is 2. Not very effective. This value suggests that the enzyme prefers to cleave peptide bonds with the following specificity: small R group—
35. If the total amount of enzyme (ET) is increased, Vmax will increase, because Vmax = k2[E]T. But KM = (k−1 + k2)/k1; that is, it is independent of substrate concentration. The middle graph describes this situation.
36.
|
Experimental condition |
Vmax |
KM |
|---|---|---|
|
a. Twice as much enzyme is used |
Doubles |
No change |
|
b. Half as much enzyme is used |
Half as large |
No change |
|
c. A competitive inhibitor is present |
No change |
Increases |
|
d. An uncompetitive inhibitor is present |
Decreases |
Decreases |
|
e. A pure non- |
Decreases |
No change |
37. (a)
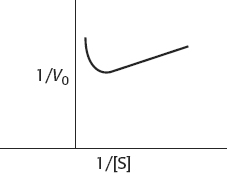
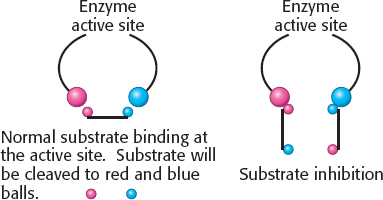
(b) This behavior is substrate inhibition: at high concentrations, the substrate forms unproductive complexes at the active site. The adjoining drawing shows what might happen. Substrate normally binds in a defined orientation, shown in the drawing as red to red and blue to blue. At high concentrations, the substrate may bind at the active site such that the proper orientation is met for each end of the molecule, but two different substrate molecules are binding.
38. The first step will be the rate-
39. The fluorescence spectroscopy reveals the existence of an enzyme–
40. (a) When [S+] is much greater than the value of KM, pH will have a negligible effect on the enzyme because S+ will interact with E− as soon as the enzyme becomes available.
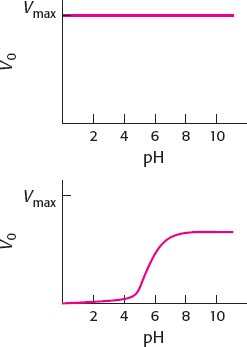
(b) When [S+] is much less than the value of KM, the plot of V0 versus pH becomes essentially a titration curve for the ionizable groups, with enzyme activity being the titration marker. At low pH, the high concentration of H+ will keep the enzyme in the EH form and inactive. As the pH rises, more and more of the enzyme will be in the E− form and active. At high pH (low H+), all of the enzyme is E−.
(c) The midpoint on this curve will be the pKa of the ionizable group, which is stated to be pH 6.
41. (a) Incubating the enzyme at 37°C leads to a denaturation of enzyme structure and a loss of activity. For this reason, most enzymes must be kept cool if they are not actively catalyzing their reactions.
(b) The coenzyme apparently helps to stabilize enzyme structure, because enzyme from PLP-
42. The slow second order step occurs when complementary sequences on two different molecules bind with one another. Once the initial sequence alignment occurs, the remainder of the molecule can quickly locate complementary sequences from this nucleation site and reanneal, a first order reaction.