30.2Aminoacyl Transfer RNA Synthetases Read the Genetic Code
Aminoacyl Transfer RNA Synthetases Read the Genetic Code
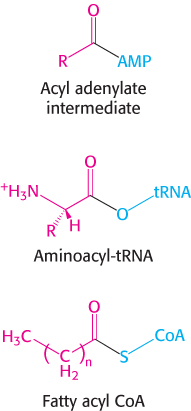
Before codon and anticodon meet, the amino acids required for protein synthesis must first be attached to specific tRNA molecules, linkages that are crucial for two reasons. First, the attachment of a given amino acid to a particular tRNA establishes the genetic code. When an amino acid has been linked to a tRNA, it will be incorporated into a growing polypeptide chain at a position dictated by the anticodon of the tRNA. Second, because the formation of a peptide bond between free amino acids is not thermodynamically favorable, the amino acid must first be activated in order for protein synthesis to proceed. The activated intermediates in protein synthesis are amino acid esters, in which the carboxyl group of an amino acid is linked to either the 2′- or the 3′-hydroxyl group of the ribose unit at the 3′ end of tRNA. An amino acid ester of tRNA is called an aminoacyl-
Amino acids are first activated by adenylation
The activation reaction is catalyzed by specific aminoacyl-

899
This activated species is a mixed anhydride in which the carboxyl group of the amino acid is linked to the phosphoryl group of AMP; hence, it is also known as aminoacyl-
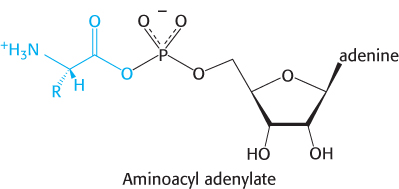
The next step is the transfer of the aminoacyl group of aminoacyl-

The sum of these activation and transfer steps is

The ΔG°′ of this reaction is close to 0, because the free energy of hydrolysis of the ester bond of aminoacyl-
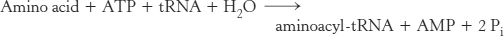
Thus, the equivalent of two molecules of ATP is consumed in the synthesis of each aminoacyl-
The activation and transfer steps for a particular amino acid are catalyzed by the same aminoacyl-
We have already encountered an acyl adenylate intermediate in fatty acid activation (Section 22.2). The major difference between these reactions is that the acceptor of the acyl group is CoA in fatty acid activation and tRNA in amino acid activation. The energetics of these biosyntheses are very similar: both are made irreversible by the hydrolysis of pyrophosphate.
Aminoacyl-tRNA synthetases have highly discriminating amino acid activation sites
Each aminoacyl-
900
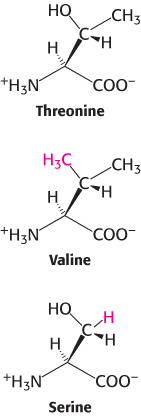
The structure of the amino acid-
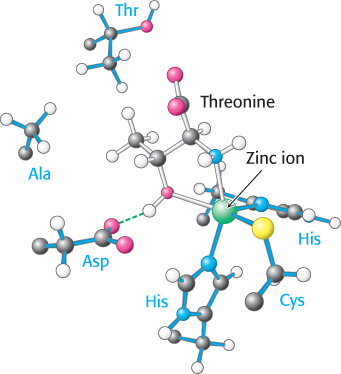
The zinc site is less able to discriminate against serine because this amino acid does have a hydroxyl group that can bind to the zinc ion. Indeed, with only this mechanism available, threonyl-
Proofreading by aminoacyl-tRNA synthetases increases the fidelity of protein synthesis
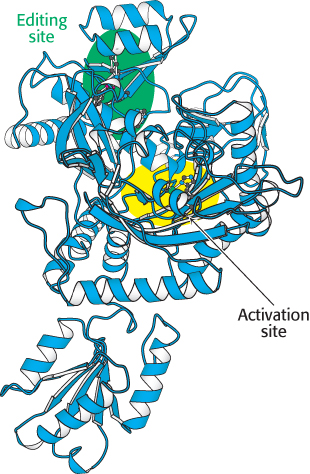
 FIGURE 30.9 Editing site. Mutagenesis studies revealed the position of the editing site (shown in green) in threonyl-
FIGURE 30.9 Editing site. Mutagenesis studies revealed the position of the editing site (shown in green) in threonyl-Threonyl-
901
Most aminoacyl-
A few synthetases achieve high accuracy without editing. For example, tyrosyl-
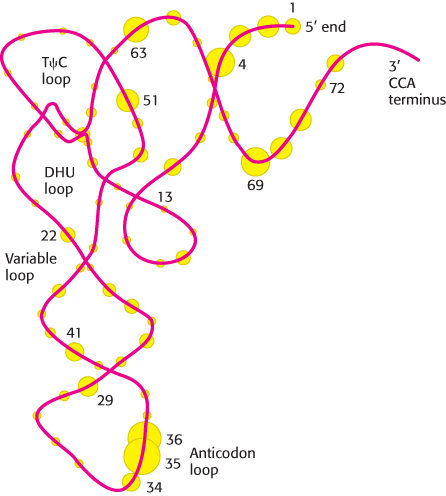
Synthetases recognize various features of transfer RNA molecules
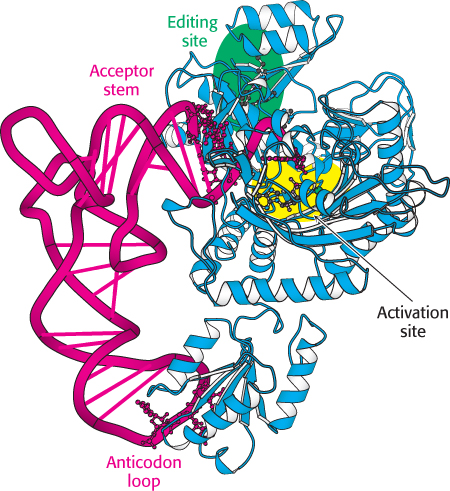
 FIGURE 30.11 Threonyl-
FIGURE 30.11 Threonyl-How do synthetases choose their tRNA partners? This enormously important step is the point at which “translation” takes place—
Some synthetases recognize their tRNA partners primarily on the basis of their anticodons, although they may also recognize other aspects of tRNA structure that vary among different tRNAs. The most direct evidence comes from crystallographic studies of complexes formed between synthetases and their cognate tRNAs. Consider, for example, the structure of the complex between threonyl-
Aminoacyl-tRNA synthetases can be divided into two classes
|
Class I |
Class II |
|---|---|
|
Arg (α) |
Ala (α4) |
|
Cys (α) |
Asn (α2) |
|
Gln (α) |
Asp (α2) |
|
Glu (α) |
Gly (α2β2) |
|
IIe (α) |
His (α2) |
|
Leu (α) |
Lys (α2) |
|
Met (α) |
Phe (α2β2) |
|
Trp (α2) |
Ser (α2) |
|
Tyr (α2) |
Pro (α2) |
|
Val (α) |
Thr (α2) |
 At least one aminoacyl-
At least one aminoacyl-
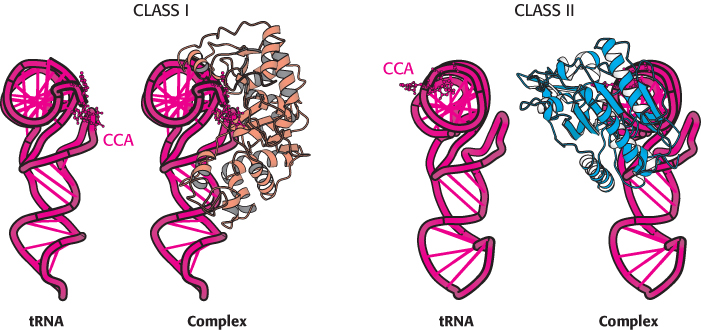
 FIGURE 30.13 Classes of aminoacyl-
FIGURE 30.13 Classes of aminoacyl-902
1. Class I enzymes acylate the 2′-hydroxyl group of the terminal adenosine of tRNA, whereas class II enzymes (except the enzyme for Phe-
2. The two classes bind ATP in different conformations.
3. Most class I enzymes are monomeric, whereas most class II enzymes are dimeric.
Why did two distinct classes of aminoacyl-