32.1Eukaryotic DNA Is Organized into Chromatin
Eukaryotic DNA Is Organized into Chromatin
Eukaryotic DNA is tightly bound to a group of small basic proteins called histones. In fact, histones constitute half the mass of a eukaryotic chromosome. The entire complex of a cell’s DNA and associated protein is called chromatin. Chromatin compacts and organizes eukaryotic DNA and its presence has dramatic consequences for gene regulation.
Nucleosomes are complexes of DNA and histones
Chromatin is made up of repeating units, each containing 200 bp of DNA and two copies each of four histone proteins H2A, H2B, H3, and H4. Histones have strikingly basic properties because a quarter of the residues in each histone are either arginine or lysine, positively charged amino acids that strongly interact with the negatively charged DNA. The protein complex is called the histone octamer. The repeating units of the histone octamer and the associated DNA are known as nucleosomes. Chromatin viewed with the electron microscope has the appearance of beads on a string (Figure 32.2); each bead has a diameter of approximately 100 Å. Partial digestion of chromatin with DNase yields the isolated beads. These particles consist of fragments of DNA about 200 bp in length bound to the histone octamer. More-
DNA wraps around histone octamers to form nucleosomes
The overall structure of the nucleosome was revealed through electron microscopic and x-
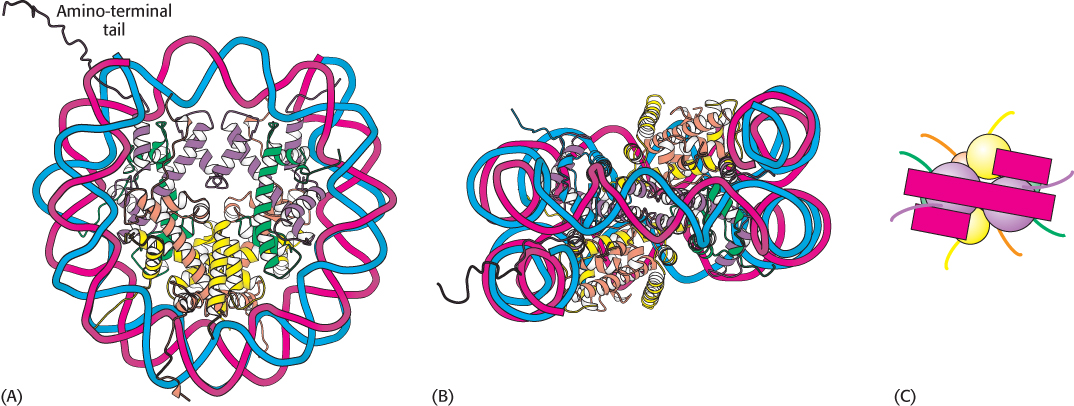
 FIGURE 32.3 Nucleosome core particle. The structure consists of a core of eight histone proteins surrounded by DNA. (A) A view showing the DNA wrapping around the histone core. (B) A 90-
FIGURE 32.3 Nucleosome core particle. The structure consists of a core of eight histone proteins surrounded by DNA. (A) A view showing the DNA wrapping around the histone core. (B) A 90-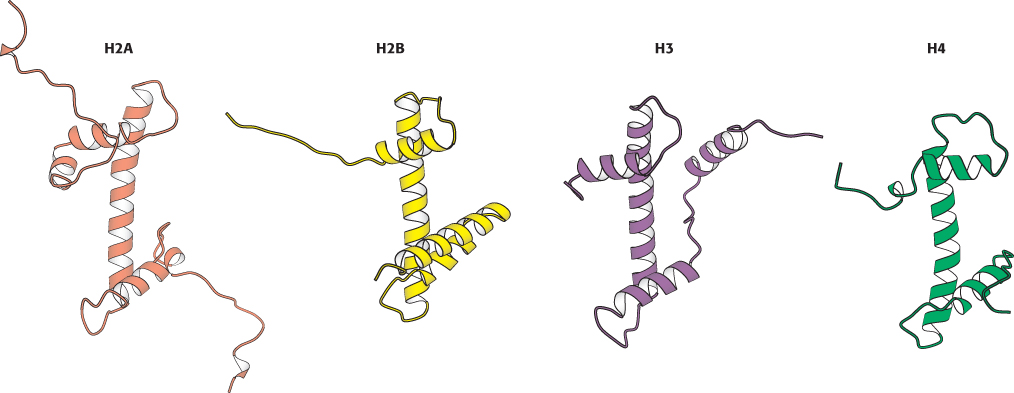
 FIGURE 32.4 Homologous histones. Histones H2A, H2B, H3, and H4 adopt a similar three-
FIGURE 32.4 Homologous histones. Histones H2A, H2B, H3, and H4 adopt a similar three-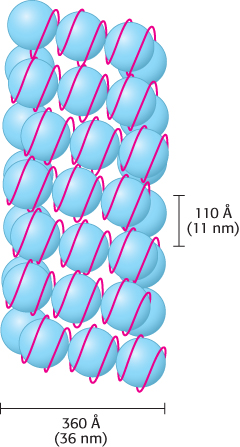
944
The DNA forms a left-
The winding of DNA around the nucleosome core contributes to the packing of DNA by decreasing its linear extent. An extended 200-
The wrapping of DNA around the histone octamer as a left-
945