33.2Taste Is a Combination of Senses That Function by Different Mechanisms
Taste Is a Combination of Senses That Function by Different Mechanisms
The inability to taste food is a common complaint when nasal congestion reduces the sense of smell. Thus, smell greatly augments our sense of taste (also known as gustation), and taste is, in many ways, the sister sense to olfaction. Nevertheless, the two senses differ from each other in several important ways. First, we are able to sense several classes of compounds by taste that we are unable to detect by smell; salt and sugar have very little odor, yet they are primary stimuli of the gustatory system. Second, whereas we are able to discriminate thousands of odorants, discrimination by taste is much more modest. Five primary tastes are perceived: bitter, sweet, sour, salty, and umami (the taste of glutamate and aspartate from the Japanese word for “deliciousness”). These five tastes serve to classify compounds into potentially nutritive and beneficial (sweet, salty, umami) or potentially harmful or toxic (bitter, sour). Tastants (the molecules sensed by taste) are quite distinct for the different groups (Figure 33.10).
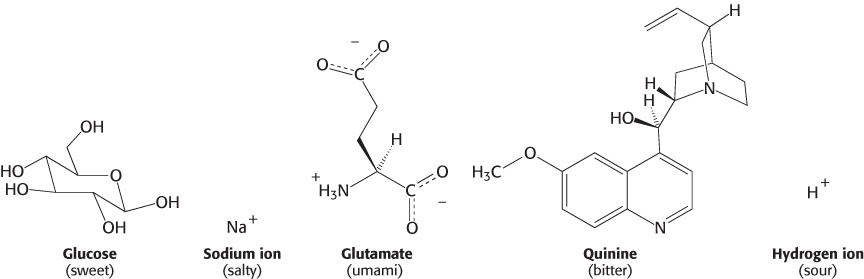
The simplest tastant, the hydrogen ion, is perceived as sour. Other simple ions, particularly sodium ion, are perceived as salty. The taste called umami is evoked by the amino acids glutamate and aspartate, the former often encountered as the flavor enhancer monosodium glutamate (MSG). In contrast, tastants perceived as sweet and, particularly, bitter are extremely diverse. Many bitter compounds are alkaloids or other plant products, many of which are toxic. However, they do not have any common structural elements or other common properties. Carbohydrates such as glucose and sucrose are perceived as sweet, as are other compounds including some simple peptide derivatives, such as aspartame, and even some proteins.
967
These differences in specificity among the five tastes are due to differences in their underlying biochemical mechanisms. The sense of taste is, in fact, a number of independent senses all utilizing the same organ, the tongue, for their expression.
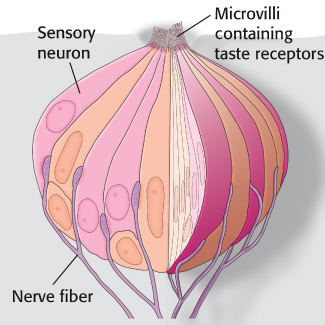
Tastants are detected by specialized structures called taste buds, which contain approximately 150 cells, including sensory neurons (Figure 33.11). Fingerlike projections called microvilli, which are rich in taste receptors, project from one end of each sensory neuron to the surface of the tongue. Nerve fibers at the opposite end of each neuron carry electrical impulses to the brain in response to stimulation by tastants. Structures called taste papillae contain numerous taste buds.
Sequencing of the human genome led to the discovery of a large family of 7TM bitter receptors
Just as in olfaction, a number of clues pointed to the involvement of G proteins and, hence, 7TM receptors in the detection of bitter and sweet tastes. The evidence included the isolation of a specific G-
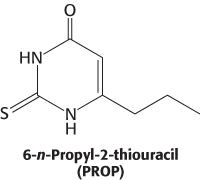
This observation suggested that this region might encode a 7TM receptor that responded to PROP. Approximately 450 kilobases in this region had been sequenced early in the human genome project. This sequence was searched by computer for potential 7TM-
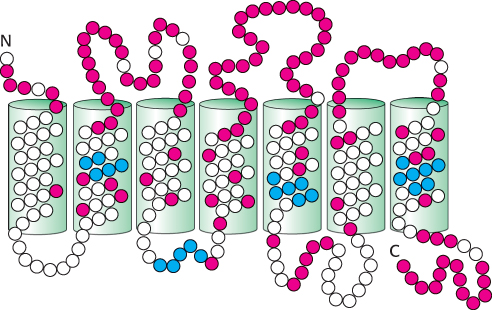
Are these proteins, in fact, bitter receptors? Several lines of evidence suggest that they are. First, their genes are expressed in taste-
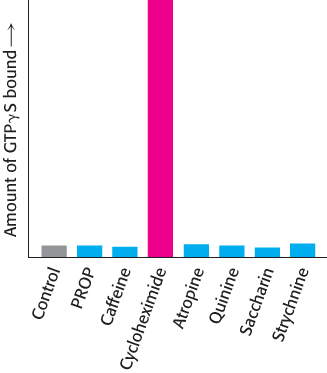
968
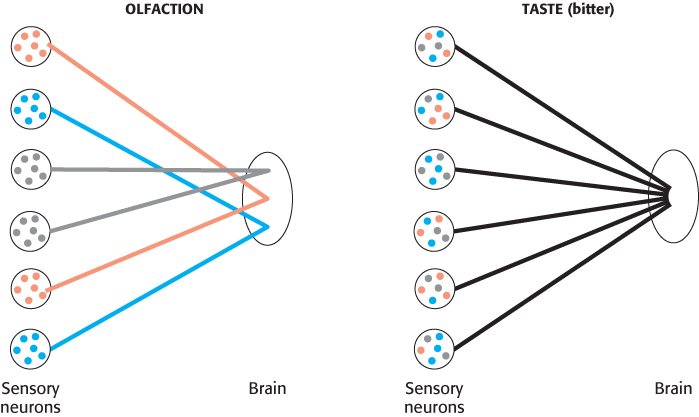
Importantly, each taste-
A heterodimeric 7TM receptor responds to sweet compounds
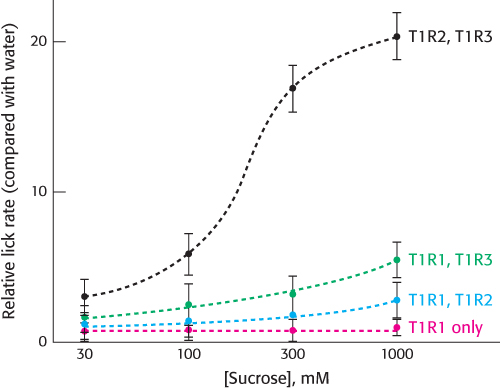
Most sweet compounds are carbohydrates, energy rich and easily digestible. Some noncarbohydrate compounds such as saccharin and aspartame also taste sweet. Members of a second family of 7TM receptors are expressed in taste-
969
The requirement for an oligomeric 7TM receptor for a fully functional response is surprising, considering our previous understanding of 7TM receptors. This discovery has at least two possible explanations. First, the sweet receptor could be a member of a small subset of the 7TM-
Umami, the taste of glutamate and aspartate, is mediated by a heterodimeric receptor related to the sweet receptor
The family of receptors responsible for detecting sweetness is also responsible for detecting amino acids. In human beings, only glutamate and aspartate elicit a taste response. Studies similar to those for the sweet receptor revealed that the umami receptor consists of T1R1 and T1R3. Thus, this receptor has one subunit (T1R3) in common with the sweet receptor but has an additional subunit (T1R1) that does not participate in the sweet response. This conclusion is supported by the observation that mice in which the gene for T1R1 is disrupted do not respond to aspartate but do respond normally to sweet tastants; mice having disrupted genes for both T1R1 and T1R3 respond poorly to both umami and sweet tastants.
Salty tastes are detected primarily by the passage of sodium ions through channels
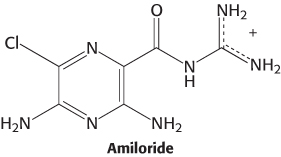
Salty tastants are not detected by 7TM receptors. Rather, they are detected directly by their passage through ion channels expressed on the surface of cells in the tongue. Evidence for the role of these ion channels comes from examining known properties of Na+ channels characterized in other biological contexts. One class of channels, characterized first for its role in salt reabsorption, is thought to be important in the detection of salty tastes because these channels are sensitive to the compound amiloride, which mutes the taste of salt and significantly lowers sensory-
An amiloride-
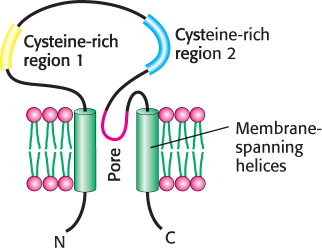
Sodium ions passing through these channels produce a significant transmembrane current. Amiloride blocks this current, accounting for its effect on taste. However, about 20% of the response to sodium remains even in the presence of amiloride, suggesting that other ion channels also contribute to salt detection.
Sour tastes arise from the effects of hydrogen ions (acids) on channels
Like salty tastes, sour tastes are detected by direct interactions with ion channels, but the incoming ions are hydrogen ions (in high concentrations) rather than sodium ions. For example, in the absence of high concentrations of sodium, hydrogen ion flow can induce substantial transmembrane currents through amiloride-
970