PROBLEMS
PROBLEMS
Question 9.1
No burst. Examination of the cleavage of the amide substrate, A, by chymotrypsin with the use of stopped-
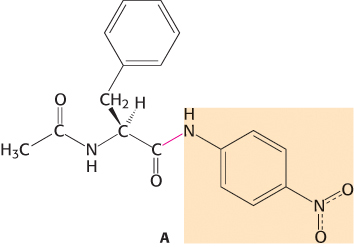
Question 9.2
Contributing to your own demise. Consider the subtilisin substrates A and B.
|
Phe- |
Phe- |
|
A |
B |
These substrates are cleaved (between Phe and X) by native subtilisin at essentially the same rate. However, the His 64-
Question 9.3
1 + 1 ≠ 2. Consider the following argument. In subtilisin, mutation of Ser 221 to Ala results in a 106-fold decrease in activity. Mutation of His 64 to Ala results in a similar 106-fold decrease. Therefore, simultaneous mutation of Ser 221 to Ala and His 64 to Ala should result in a 106 × 106 = 1012-fold reduction in activity. Is this reduction correct? Why or why not?
Question 9.4
Adding a charge. In chymotrypsin, a mutant was constructed with Ser 189, which is in the bottom of the substrate-
Question 9.5
Conditional results. In carbonic anhydrase II, mutation of the proton-
Question 9.6
How many sites? A researcher has isolated a restriction endonuclease that cleaves at only one particular 10-
Question 9.7
Is faster better? Restriction endonucleases are, in general, quite slow enzymes with typical turnover numbers of 1 s−1. Suppose that endonucleases were faster, with turnover numbers similar to those for carbonic anhydrase (106 s−1), such that they act faster than do methylases. Would this increased rate be beneficial to host cells, assuming that the fast enzymes have similar levels of specificity?
Question 9.8
Adopting a new gene. Suppose that one species of bacteria obtained one gene encoding a restriction endonuclease by horizontal gene transfer. Would you expect this acquisition to be beneficial?
Question 9.9
Chelation therapy. Treatment of carbonic anhydrase with high concentrations of the metal chelator EDTA (ethylenediaminetetraacetic acid) results in the loss of enzyme activity. Propose an explanation.
Question 9.10
An aldehyde inhibitor. Elastase is specifically inhibited by an aldehyde derivative of one of its substrates:
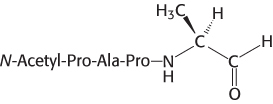
Which residue in the active site of elastase is most likely to form a covalent bond with this aldehyde?
What type of covalent link would be formed?
284
Question 9.11
Identify the enzyme. Consider the structure of molecule A. Which enzyme discussed in the chapter do you think molecule A will most effectively inhibit?
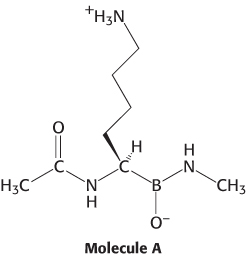
Question 9.12
Acid test. At pH 7.0, carbonic anhydrase exhibits a kcat of 600,000 s−1. Estimate the value expected for kcat at pH 6.0.
Question 9.13
Restriction. To terminate a reaction in which a restriction enzyme cleaves DNA, researchers often add high concentrations of the metal chelator EDTA (ethylenediaminetetraacetic acid). Why does the addition of EDTA terminate the reaction?
Question 9.14
Labeling strategy. ATP is added to the myosin ATPase domain in water labeled with18 O. After 50% of the ATP has been hydrolyzed, the remaining ATP is isolated and found to contain18O. Explain.
Question 9.15
Viva la resistance. Many patients become resistant to HIV protease inhibitors with the passage of time owing to mutations in the HIV gene that encodes the protease. Mutations are not found in the aspartate residue that interacts with the drugs. Why not?
Question 9.16
More than one way to skin kcat. Serine 236 in Dictyostelium discoideum myosin has been mutated to alanine. The mutated protein showed modestly reduced ATPase activity. Analysis of the crystal structure of the mutated protein revealed that a water molecule occupied the position of the hydroxyl group of the serine residue in the wild-
Question 9.17
A power struggle. The catalytic power of an enzyme can be defined as the ratio of the rate of the enzyme catalyzed reaction to that for the uncatalyzed reaction. Using the information in Figure 9.15 for subtilisin and in Figure 9.22 for carbonic anhydrase, calculate the catalytic powers for these two enzymes.
Question 9.18
Wounded but not dead. How much activity (in terms of relative kcat values) does the version of subtilisin with all three residues in the catalytic triad mutated have compared to uncatalyzed reaction? Propose an explanation.
Mechanism Problem
Question 9.19
Complete the mechanism. On the basis of the information provided in Figure 9.17, complete the mechanisms for peptide-