Bread Mold, Neurospora crassa
The bread mold, Neurospora crassa, is a filamentous ascomycete fungus. It was one of the first eukaryotic microorganisms adopted for genetic studies. About 75% of all fungi are ascomycetes, and the remainder are basidiomycetes (including mushrooms). About 90% of ascomycetes are filamentous (and often with multinucleate hyphae, as further explained below), and the rest are single-
The war between filamentous fungi and bacteria has yielded important antibiotics, including penicillin. These complex organic molecules are found nowhere else in nature, and accordingly, more than 40% of Neurospora genes have no identifiable counterpart in any other organism studied. The ecological niche filled by filamentous fungi includes the decay of many types of biological material. This may explain why Neurospora can grow on highly unusual carbon and nitrogen sources—
A unique feature of Neurospora is the arrangement in the spore case (ascus) of the spores produced from meiosis of a single diploid nucleus. The spores are linearly arranged in the order in which they are produced by the meiotic divisions. With the ability to precisely separate spores and study them individually, researchers have an ideal model for the analysis of meiotic recombination. Neurospora is also attractive for studying several gene silencing mechanisms, some of which also exist in multicellular eukaryotes.
Early Studies of Neurospora as a Model Organism
Neurospora achieved its early standing as a model organism because of its haploid state, rapid growth, production of millions of asexual spores per colony, and ease of culture on simple, defined media. The capacity to synthesize all its essential biomolecules from media with known ingredients propelled Neurospora into the spotlight in the 1940s as an ideal model in which to study the genetics of metabolic pathways that are similar in all cells. Of particular historical note was the use of Neurospora by George Beadle and Edward Tatum in studies that formed the basis for the “one gene, one polypeptide” hypothesis. This work ushered in a new era of molecular genetics and the use of microorganisms in genetic studies (see Figure 2-23).
Life Cycle
The asexual life cycle of Neurospora starts with haploid spores released from microconidia and macroconidia (Figure A-
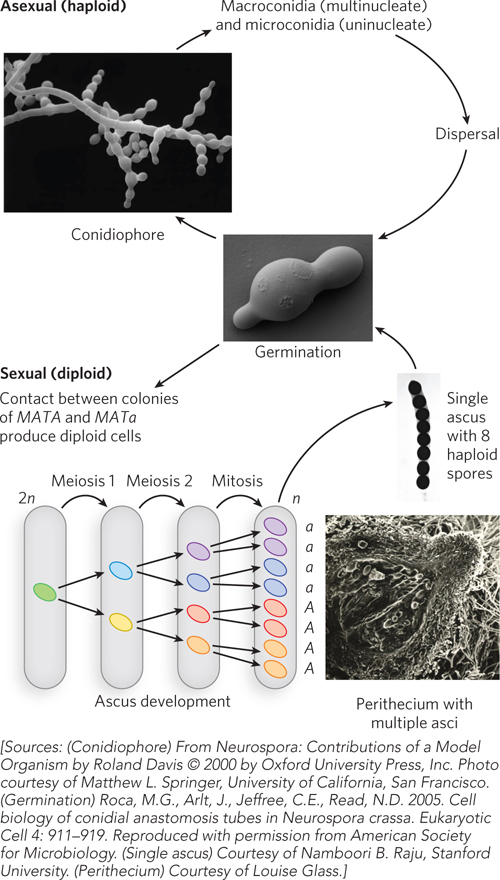
Two different alleles at the mating type locus, mat A and mat a, control the Neurospora sexual cycle. Spores contain either the MATA or MATa allele. Neurospora is hermaphroditic, and both mating types produce female reproductive structures (protoperithecia). However, fertilization can occur only between reproductive structures of opposite mating type (protoperithecia and conidia/microconidia of opposite mating type). Fertilization results in formation of hyphae containing both mating type nuclei (a dikaryon). Nuclear fusion in the dikaryon is followed by meiotic cell divisions, each diploid nucleus yielding four haploid spores (ascospores) in an ascus (see Figure A-
A-
Genetic Techniques
Mutagenesis Neurospora asexual spores can be mutagenized by treatment with ionizing irradiation. Irradiated spores are germinated in complete media and allowed to sporulate, forming stocks of mutant lines. Spores from these lines can then be screened for phenotypes of interest, and mutations can be mapped by genetic crosses.
Introduction of DNA Neurospora readily takes up exogenous DNA by transformation. The DNA must integrate into the genome to be stably inherited. The antibiotic hygromycin can be used to select for Neurospora with a transgene. Unlike in yeast, DNA insertion rarely occurs through homologous recombination in Neurospora; insertion is more often random and untargeted.
Gene Knockouts There is an unusual way of generating a specific gene knockout in Neurospora. A cell with a duplicate gene (constructed either by transformation or by crosses using a translocation strain), when crossed, undergoes a process called RIP (repeat-
Sporulation As Neurospora chromosomes segregate during meiosis, the daughter cells become arranged linearly in a long axis. The frequency of crossing over can be used to map the distance of a mutant allele locus from the centromere.
Neurospora as a Model Organism Today
Meiotic Recombination As we have seen, Neurospora packages its ascospores from a single meiotic event in an arrangement (in the ascus) that reflects the order of chromosomal segregation during meiosis. This feature makes Neurospora an attractive system for studies of crossover recombination during meiosis.
Circadian Rhythms Neurospora has a daily circadian rhythm of conidial spore formation. Conidial spores are conspicuous to the naked eye, so the phenotype of rhythmic behavior can be examined on Petri plates or in race tubes—
Silencing Mechanisms Neurospora contains at least three different silencing mechanisms that are thought to have evolved as genomic defenses against invading DNA, such as transposons and viruses. Meiotic silencing by unpaired DNA (MSUD) is a process in which any unpaired gene that is detected during meiosis is silenced. Quelling occurs in haploid cells and detects duplicate sequences. RIP detects duplicated sequences just before meiosis, resulting in methylation and G≡C-
A-