DATA ANALYSIS PROBLEM
Nishimura, S., D.S. Jones, E. Ohtsuka, H. Hayatsu, T.M. Jacob, and H.G. Khorana. 1965. Studies on polynucleotides XLVII: The in vitro synthesis of homopeptides as directed by a ribopolynucleotide containing a repeating trinucleotide sequence—
Question 17.18
Once researchers had developed a few key strategies, the genetic code was solved within just a few years in the mid-
Using methods developed in the Khorana laboratory, Nishimura and colleagues examined the polypeptides generated by an oligonucleotide consisting of AAG repeats. In the first experiment (Figure 1), they examined the binding of radioactively labeled aminoacyl-
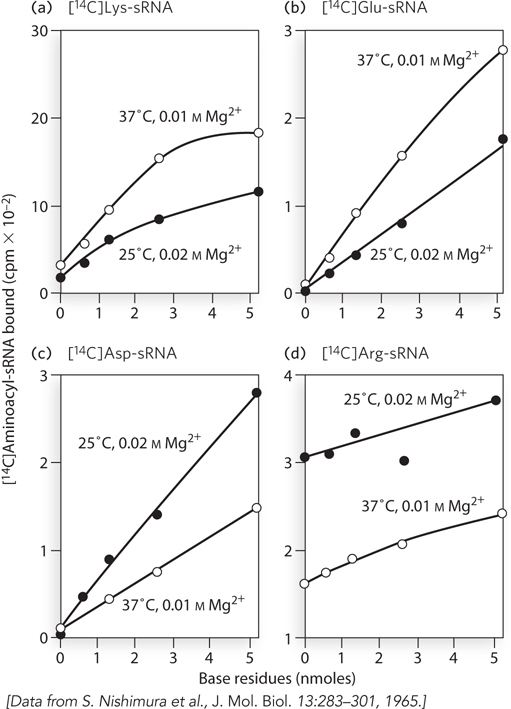
615
Given our present understanding of the genetic code, what is the maximum number of different labeled aminoacyl-
tRNAs that could be bound to the ribosome in response to this oligonucleotide? Given the tenuous understanding of the code in 1965, can you suggest an explanation for the positive results obtained with the four different tRNAs in this experiment?
One of the possible codons present in the (AAG)n oligonucleotide was assigned to lysine, based on a series of experiments including the one shown in Figure 2. The researchers used the method advanced by Nirenberg to determine which trinucleotide would stimulate binding of [14C]Lys-
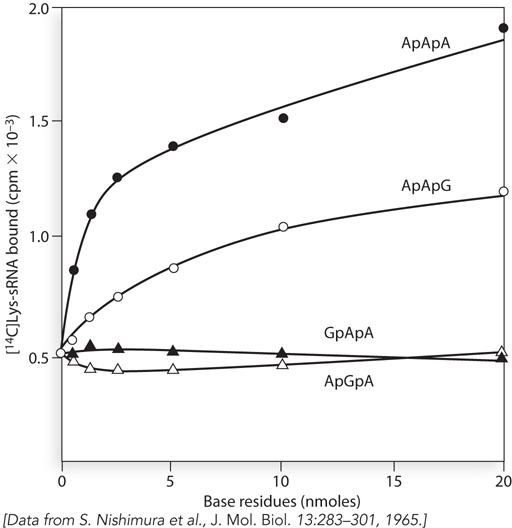
Which codon did the researchers assign to lysine?
The researchers also tested the trinucleotide AAA for its response to [14C]Lys-
tRNALys (see Figure 2). Suggest why they would include AAA in their experiment, even though AAA was not represented in the (AAG)n repeating oligonucleotide.
Next, the researchers turned their attention to the tRNAs for glutamine. With [14C]Glu-
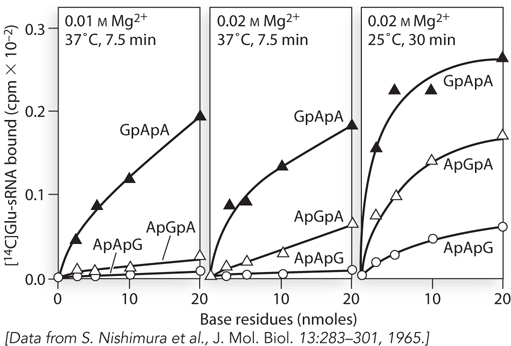
From these results, which codon did the authors assign to Glu?
Suggest why all three codons elicited a positive response in the experiment shown in the rightmost panel of Figure 3.
In their tests, the researchers then examined the production of homopolymers of amino acids in response to (AAG)n. They found poly(Glu), poly(Arg), and poly(Lys), but no poly(Asp).
Based on all of the results shown here, what are the most likely codon assignments for the codons present in (AAG)n?
Go back to question (b) and suggest why Asp-
tRNAAsp bound to ribosomes in response to this oligonucleotide in the first experiment (see Figure 1).