DATA ANALYSIS PROBLEM
Brent, R., and M. Ptashne, M. 1985. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43:729–
Question 19.15
The concept that eukaryotic regulatory proteins have multiple functional domains was arrived at in stages. However, a few experiments stand out, such as the study by Roger Brent and Mark Ptashne published in 1985. When the study began, one known mechanism for activation of transcription by an activator protein was simply direct interaction with RNA polymerase. The investigators also considered an alternative mechanism: that the transcription activator functioned by altering the structure of the DNA to which it bound, facilitating the binding of RNA polymerase.
The study focused on two different regulatory proteins. The first was a well-
Suggest why the investigators did not simply examine the effects of the LexA-
Gal4 fusion protein on the GAL1 gene already in yeast cells. The investigators first tested the LexA-
Gal4 protein in E. coli cells that lacked their own LexA- encoding gene, and showed that cells containing the fusion protein repressed transcription of genes normally repressed by LexA. Why was this control experiment undertaken?
Next, they carried out a series of measurements of β-galactosidase activity with the two-
plasmid system in yeast cells, with the results shown in Table 1 (data from their published table). In the table, β-galactosidase activity is given in units of blue color produced in the conversion of substrate to product. UAS is upstream activator sequence; UASC1 and UASC2 are binding sites for activator proteins that function at the CYC1 gene; and UASG is the normal binding site for Gal4p. UASG consists of four separate binding sites for Gal4p, each 17 bp long. The 17mer is a site with just one of these sequences. The abbreviation “op” means operator, which is the LexA- binding site; −178 and −577 indicate the distance in base pairs between the operator and the transcription start site. In the yeast cells used for the study, the genes encoding the endogenous Gal4p and the CYC1 gene activators were all present and functional. 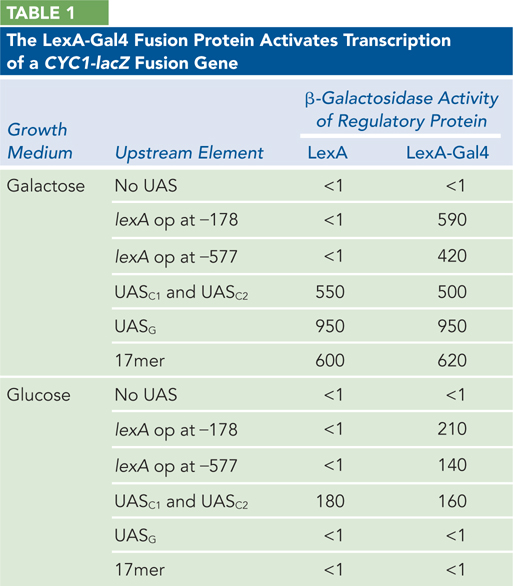
692
How effective is the LexA-
Gal4 fusion protein (acting at the LexA operator) at activating gene expression, relative to the cellular Gal4p acting at UASG? Is transcription activated by the LexA protein by itself?
Does the location of the LexA operator affect the activity of the LexA-
Gal4 fusion protein? When UASG or the 17mer is upstream from the CYC1-
lacZ reporter gene, why is expression seen only when the cells were grown in galactose?What result in Table 1 indicates that the LexA-
Gal4 fusion protein is activating transcription by direct interaction with RNA polymerase, not by altering the structure of the DNA to which the polymerase binds?