21.1 BASIC MECHANISMS OF EUKARYOTIC TRANSCRIPTIONAL ACTIVATION
As in bacteria, the basal level of eukaryotic transcription is determined by the effect of regulatory sequences on the function of RNA polymerase and its associated transcription factors. As we discussed in Chapter 19, the nature of the eukaryotic genome lends itself to different regulatory strategies from those used in bacteria. The eukaryotic genome is packaged in chromatin, which presents a physical block to RNA polymerases, and therefore the majority of eukaryotic genes are repressed in their default (ground) state and require protein activators to stimulate expression. Because of the large size of eukaryotic genomes and the need to guard against nonspecific protein-DNA interactions, the binding of multiple protein regulators is required to activate each gene. As a result, eukaryotic promoters are more complex than their bacterial counterparts and contain many more regulatory protein–binding sites. In reality, though, the additional complexity in eukaryotes is handled in strategic ways that are not as complicated as we might expect, given the overwhelming difference between a bacterium and an animal.
Eukaryotic Transcription Is Regulated by Chromatin Structure
The genomic DNA of eukaryotes wraps around small basic proteins called histones to form nucleosomes, the building blocks of chromatin (see Figure 10-4). The transcription machinery must necessarily deal with chromatin structure in order to access particular genes. As a result, eukaryotic genes are generally expressed at low levels—or not at all—in the absence of regulatory proteins.
Chromatin structure is controlled and altered by at least three interrelated mechanisms: ATP-dependent changes in nucleosome positioning on the DNA, posttranslational chemical modifications of histone proteins, and substitution of specialized histone variants into chromatin. These mechanisms were discussed in detail in Chapter 10, and we recap briefly here. Nucleosome remodeling complexes use ATP to shift nucleosomes along the DNA. Active promoters contain open regions with nucleosomes positioned away from the promoter region, allowing access to transcription factors. Some posttranslational modifications of histones, including acetylation by histone acetyltransferases (HATs), result in the decondensing of chromatin and provide access to DNA-binding factors; proteins containing a bromodomain bind acetylated histones and facilitate opening of the chromatin structure. Alternatively, histone modifications cause chromatin to become tightly closed to transcription. For example, methylated histones are bound by proteins containing chromodomains, and these proteins help condense the chromatin. Chromatin structure is also modulated by several histone variants. These proteins are homologous to the common histones and can take their place in nucleosomes, but they also contain amino acid extensions that have a variety of functional consequences.
In the eukaryotic cell cycle, interphase chromosomes appear to be dispersed and amorphous. However, chromosomes are not uniform structures, and several different forms of chromatin can be found along each chromosome. About 10% of the chromatin in a typical eukaryotic cell is in a much more condensed form than the rest of the chromatin. This form, heterochromatin, is transcriptionally inactive. Although heterochromatin does not contain any genes (which is why it is inactive), the heterochromatin structure itself is known to be repressive, because, experimentally, a gene is silenced when it is placed in heterochromatin. Heterochromatin is often associated with particular chromosome structures, including centromeres and telomeres. The remaining, less condensed chromatin is called euchromatin (Figure 21-1).

Figure 21-1: Heterochromatin and euchromatin. Nucleosomes in heterochromatin are tightly packed together, and the DNA is transcriptionally silent. Nucleosomes in euchromatin are spaced farther apart, and the DNA can be decondensed by the loss of histone H1, thus becoming accessible to the transcription machinery. The two chromatin states are regulated by histone modifications (represented by red asterisks) and the binding of other factors (see Chapter 10).
Transcription of a eukaryotic gene is strongly repressed when its DNA is condensed within heterochromatin, but in euchromatin, some of the DNA is transcriptionally active. Regions of transcriptionally active DNA can be detected on the basis of their increased sensitivity to nuclease-mediated degradation. Nucleases such as DNase I tend to cleave the DNA of carefully isolated chromatin into fragments of multiples of about 200 bp, reflecting the regular repeating structure of the nucleosome (see Figure 10-1). However, in actively transcribed regions, the fragments produced by nuclease activity are smaller and more heterogeneous in size. Actively transcribed regions contain hypersensitive sites, sequences especially sensitive to DNase I, which are typically found in noncoding regions within 1,000 bp of the 5′ ends of transcribed genes. In some genes, hypersensitive sites are found farther from the 5′ end, or near the 3′ end, or even within the gene itself. The presence of hypersensitive sites suggests that DNA in that region is not packaged in the regular repeating nucleosomal structure.
Many hypersensitive sites correspond to binding sites for known regulatory proteins, and the relative absence of nucleosomes in these regions may allow the binding of these proteins. Nucleosomes are entirely absent in some regions that are very active in transcription, such as the rRNA genes. Transcriptionally active chromatin also tends to be deficient in histone H1, which binds the linker DNA between nucleosome particles.
Histones within transcriptionally active chromatin and heterochromatin also differ in their patterns of covalent modification. The C-terminal tails of the core histones are modified by the acetylation and methylation of Lys and Arg residues, phosphorylation of Ser or Thr residues, and ubiquitination or sumoylation (see Chapter 22). In particular, the acetylation-deacetylation of histones figures prominently in the processes that activate chromatin for transcription. The HAT-mediated acetylation of multiple Lys residues in the N-terminal domains of histones H3 and H4 can reduce the affinity of the entire nucleosome for DNA. Acetylation may also prevent or promote interactions with other proteins involved in regulating transcription. When transcription of a gene is no longer required, acetylation of nucleosomes in that vicinity is reduced by the activity of histone deacetylases (HDACs), resulting in condensation of the chromatin to reduce or inactivate gene transcription. HDACs often function through protein-protein interactions, such as by binding corepressors or acting as components of chromatin remodeling complexes.
A general model for deacetylation and gene inactivation is shown in Figure 21-2. In addition to the removal of certain acetyl groups, new covalent modifications of histones mark chromatin as transcriptionally inactive. For example, the Lys residue at position 9 in histone H3 is often methylated in heterochromatin.
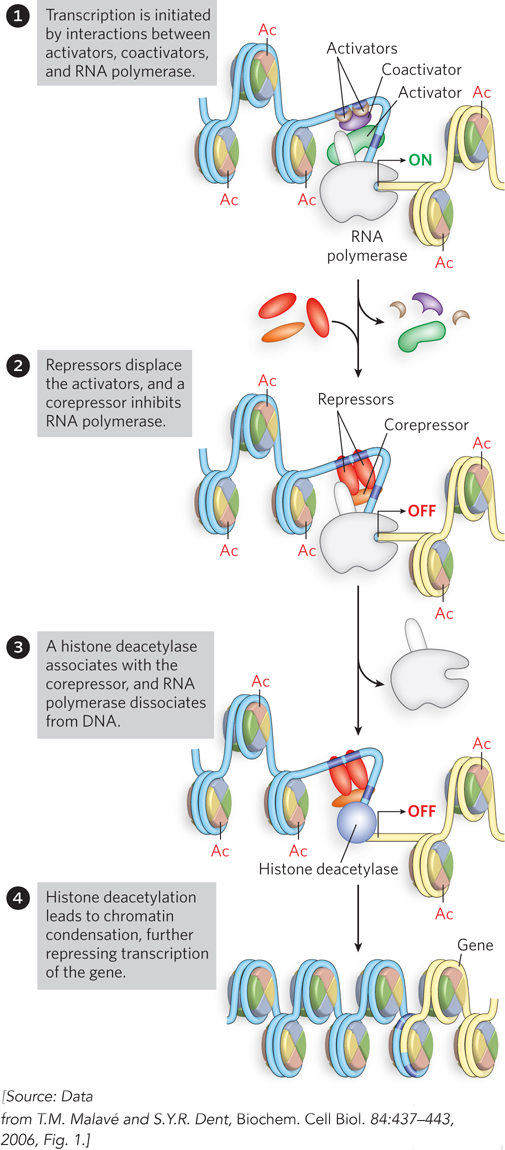
Figure 21-2: Gene inactivation by histone deacetylation. Removal of acetyl groups from histones leads to dissociation of RNA polymerase and condensation of the chromatin.
Gene regulation through histone modifications is typically achieved through activation or inhibition of transcription initiation. However, an exciting finding has demonstrated that chromatin structure is also involved in the control of mRNA splicing for some genes (see this chapter’s Moment of Discovery). This may seem surprising, given that histones do not bind RNA. Yet Gcn5, a well-studied transcription factor with HAT activity, is now known to also affect RNA processing: loss of Gcn5 HAT activity prevents proper pre-mRNA splicing in yeast, because components of the splicing machinery fail to properly bind the pre-mRNA splice sites (Highlight 21-1). This example shows how different types of regulation (in this case, transcription and mRNA processing) can be interrelated. It seems likely that processes previously thought to be isolated and separable events may be interwoven in complex regulatory networks in the living cell.
Positive Regulation of Eukaryotic Promoters Involves Multiple Protein Activators
Each of the three eukaryotic RNA polymerases has little or no intrinsic affinity for its promoters. Instead, initiation of transcription almost always requires activator proteins. An important reason for the apparent predominance of positive regulation is clear from the earlier discussion: chromatin structure effectively renders most promoters inaccessible, so genes are normally silent in the absence of other regulation. The structure of chromatin affects access to some promoters more than others, but repressor binding to DNA to block access of RNA polymerase (negative regulation) would often be simply redundant. Other factors are also at play in the use of positive regulation, however, and speculation generally centers on two: the large size of eukaryotic genomes and the greater efficiency of positive regulation.
Because eukaryotes have much larger genomes than bacteria, there is an increased likelihood that a specific binding sequence for a regulatory protein will occur randomly in other regions of the DNA. Recall that a sequence of n nucleotides will occur randomly every 4n bp. Thus, any single regulatory protein with a small binding site will probably bind nonspecifically to multiple places in the eukaryotic genome (see the How We Know section at the end of this chapter). Specific transcriptional activation of a gene through the binding of one regulatory protein to a small binding site, as often occurs in bacteria, would be ineffective in eukaryotes. In theory, specificity of regulator binding would be increased if the DNA sequences recognized by the proteins were longer. Yet eukaryotes did not evolve in this way: eukaryotic regulators do not bind longer DNA sequences than their bacterial counterparts. Instead, to achieve specific transcriptional activation of a gene, eukaryotes employ multiple regulatory proteins, or transcription factors, each of which binds a short sequence; successful gene activation occurs only when all the factors are bound at their individual sites. This “combination” of factors for activating one gene is used in combinatorial control (see Section 21.2).
To accommodate the binding of multiple transcription factors, eukaryotic promoters are necessarily more complicated than their bacterial counterparts (Figure 21-3). Take, for example, a typical promoter recognized by RNA polymerase II (Pol II), the enzyme responsible for mRNA synthesis. Many (but not all) Pol II promoters include the TATA box and Inr (initiator) sequences, with their standard spacing (see Figure 15-20). These sequences comprise the core promoter.
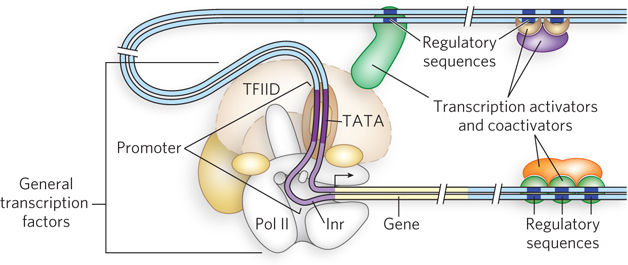
Figure 21-3: A typical eukaryotic promoter. General transcription factors and RNA polymerase II bind the promoter, assisted by transcription activators. Activator-binding sites (regulatory sequences) can be distant from the promoter and located either before or after the gene. Activators bind regulatory sequences in DNA directly, whereas coactivators bind activators instead of DNA. Activation of Pol II is mediated by coactivators binding to core subunits of the polymerase through DNA looping.
Eukaryotic genes also include regulatory sequences called enhancers in higher eukaryotes and upstream activator sequences (UASs) in yeast, to which transcription activators bind. These sequences cannot all be positioned adjacent to the promoter—there is simply not enough room to accommodate the binding of so many regulatory proteins. The binding sites for multiple transcription factors must be able to act at a distance. In fact, they can be surprisingly far from the promoter. A typical enhancer may be hundreds or even thousands of base pairs upstream from the transcription start site, or downstream from the gene, or even within the gene itself. When bound by the appropriate regulatory proteins, an enhancer increases transcription at nearby promoters, regardless of its orientation in the DNA. Yeast UASs function in a similar way, although generally they must be positioned upstream and within a few hundred base pairs of the transcription start site. An average Pol II promoter may be affected by half a dozen regulatory sequences of this type, and many promoters are even more complex. In contrast, bacteria have very few genes that use a distantly bound transcription activator.
The more complex the eukaryotic organism, the more complex its promoters are likely to be. For example, mammalian promoters are generally much more complex than yeast promoters (Figure 21-4).
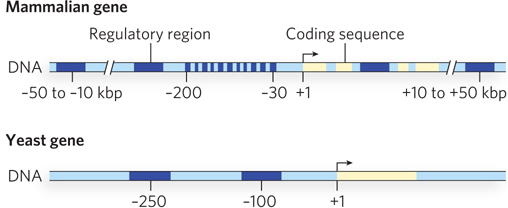
Figure 21-4: A comparison of mammalian and yeast promoter regions. The promoter regions of multicellular organisms, such as mammals, contain more control elements than those of unicellular eukaryotes, such as yeast. This reflects the need in higher eukaryotes for changes in gene expression during development and for intercellular communication. All regulatory regions are shown in dark blue, coding regions in yellow.
HIGHLIGHT 21-1 A CLOSER LOOK: The Intertwining of Transcription and mRNA Splicing
Initiation is the most highly regulated step in transcription, an intricate process that requires, in eukaryotes, the coordinated action of numerous proteins. Transcription generates a pre-mRNA needing many modifications before it can be transported to the cytoplasm for translation. One of the most complex modifications en route to active mRNA is the removal of introns. The splicing machinery requires more than 100 proteins and five different splicing RNAs with complicated three-dimensional structures. Splicing is generally regarded as a separate step occurring after transcription initiation, or even after generation of the entire pre-mRNA, partly because of the complexity of the transcription and splicing processes. So it was surprising to discover that these two complicated processes—transcription and splicing—can happen simultaneously for some genes: transcription seems to deposit the U2 snRNP component of the spliceosome at specific sites in the pre-mRNA as it is synthesized. These sites correspond to branch points, the sites containing the 2′-OH nucleophile that initiates intron splicing and results in a branched, lariat-type structure when the intron RNA is excised (see Chapter 16). To explain why transcription and splicing would coordinate in this fashion, researchers have proposed that the rate of transcription elongation may be regulated by the spliceosome to help pick and choose alternative splice sites, thereby controlling the relative levels of different mRNAs produced from a pre-mRNA.
The true picture of what is going on is even more complicated, however, as revealed by recent work in Tracy Johnson’s laboratory (see this chapter’s Moment of Discovery). Johnson made the fascinating observation that Gcn5, a histone acetyltransferase (HAT), is an integral component in the coregulation of transcription and pre-mRNA splicing. The HAT activity of Gcn5, like other HATs, can alter chromatin structure, which is thought to be important in regulating transcription initiation. But results from Johnson’s lab demonstrate that accurate mRNA splicing, too, requires the Gcn5 HAT activity.
How does a HAT help splicing? After all, RNA is not bound by histones, so what role does Gcn5 play in the splicing process? Johnson found that the coordination between transcription and splicing occurs even before the pre-mRNA is fully synthesized. The first evidence hinting at this conclusion came from genetic experiments showing that deletion of the gene encoding Gcn5 (and not other yeast lysine acetyltransferases that target histones) is lethal in yeast cells that also lack either of the genes encoding the U2 snRNP proteins Lea1 and Msl1. Neither Lea1 nor Msl1 is an essential protein in yeast, except when Gcn5 is missing.
Next, using the technique of chromatin immunoprecipitation (ChIP), Johnson’s group showed that spliceosomal proteins are recruited directly to an intron branch point within the well-characterized DBP2 gene. In the ChIP experiment, individual snRNP particles are formaldehyde cross-linked to the transcription complex or to the nascent RNA and immunoprecipitated (see Figure 10-21). When the associated DNA is amplified using specific PCR primer sets, the signal is enriched in regions of the gene where the snRNPs associate with the corresponding pre-mRNA. Johnson’s results revealed that antibodies to Lea1, a component of the spliceosome, immunoprecipitated a relatively large amount of DNA corresponding to the branch point of the DBP2 pre-mRNA (Figure 1a, b). Recruitment of Lea1 to the branch point depended on the presence and catalytic activity of Gcn5 (Figure 1c). A control experiment showed that the occupancy by Pol II of these regions of the DPB2 gene was unaltered, whether or not Gcn5 was active (Figure 1d). Thus, the data indicate that Gcn5 sets the stage for the recruitment of spliceosomal components before the splice site junctions are even transcribed. Further experiments have demonstrated the same results for other genes.
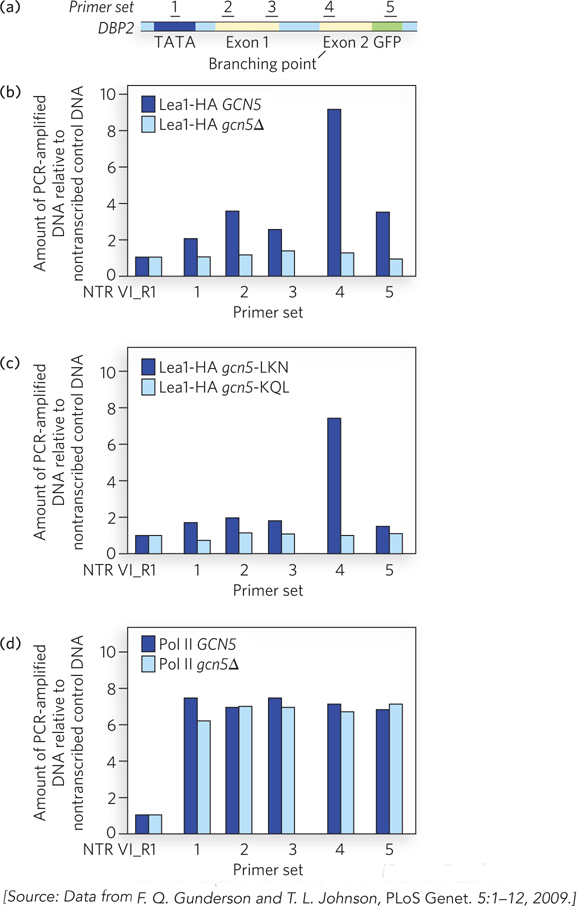
FIGURE 1 Gcn5 activity helps recruit spliceosomal components to DBP2 pre-mRNA. (a) Numbers represent regions of DNA in the DBP2 gene that are amplified in the ChIP analysis. (b) ChIP analysis of yeast cells expressing an engineered version of Lea1 tagged with a hemagglutinin (HA) peptide. Lea1-HA was immunoprecipitated with anti-HA antibodies, and Lea1 occupancy in the indicated regions of DBP2 was compared with that of a nontranscribed region of DNA (NTR VI_R1). Sets of PCR primers corresponding to the regions indicated in (a) were used to amplify specific segments of chromatin after Lea1-HA immunoprecipitation. Dark blue bars are data for cells with wild-type Gcn (GCN5); light blue bars are data for cells with a Gcn5 deletion (gcn5∆). (c) ChIP analysis as in (b), but the dark blue bars are results for cells with a point mutation in a nonessential region of Gcn5 (gcn5-LKN) and the light blue bars are results for cells with a point mutation in the Gcn5 active site (gcn5-KQL). (d) ChIP analysis as in (b), but this control experiment uses Pol II instead of Lea1.
Johnson also proposes other explanations for these observations. One possibility is that Gcn5 acetylates nonhistone proteins, perhaps even spliceosomal subunits. Another possibility is that hyperacetylation of histones at the promoter may facilitate recruitment of the splicing apparatus. Understanding the full details of coordinated regulation of transcription initiation, histone acetylation, and recruitment of the spliceosomal machinery will take considerably more time and work.
The requirement for the binding of several transcription activators to several specific DNA sequences vastly reduces the probability of the random occurrence of a functional juxtaposition of all the necessary binding sites. In principle, a similar strategy could be used by multiple negative-regulatory elements. However, positive regulation is simply more efficient. From an energy standpoint, it makes more sense for the cell to synthesize several activators to promote transcription of the subset of genes needed at that time, rather than constantly synthesize one or more repressors for every gene in the genome to keep them turned off until needed. Positive regulation of transcription predominates in eukaryotes, although, as we will see, there are some examples of negative regulation.
To further conserve resources, differently regulated eukaryotic promoters often use some of the same protein activators, so diverse promoters can have some of the same binding sequences. However, only a specific combination of regulatory factors can unlock a given promoter and activate transcription of that gene. With this mechanism, the cell can achieve specificity of gene regulation with a smaller number of transcription activators than if each gene were regulated by a set of unique proteins (see Figure 19-14). Some regulatory proteins facilitate transcription at hundreds of promoters, whereas others are specific for only a few promoters. In addition, many transcription activators are sensitive to the binding of effector signal molecules, providing the capacity to activate or deactivate transcription in response to a changing cellular environment.
Transcription Activators and Coactivators Help Assemble General Transcription Factors
Successful binding of active Pol II holoenzyme at one of its promoters usually requires the action of three types of regulatory proteins: general transcription factors, DNA-binding transcription activators, and coactivators. General (basal) transcription factors are required at every Pol II promoter; DNA-binding transcription activators, or DNA-binding transactivators, bind to enhancers or UASs to facilitate transcription; and coactivators act indirectly—by binding other proteins rather than DNA—and are required for essential communication between the DNA-binding transactivators and the complex composed of Pol II and the general transcription factors (Figure 21-5a). Sometimes, a variety of repressor proteins can interfere with communication between Pol II and the DNA-binding transactivators, resulting in repression of transcription (Figure 21-5b). In fact, some proteins act as an activator or coactivator at one promoter and a repressor or corepressor at another promoter. Here we focus on the protein complexes shown in Figure 21-5a and how they interact to activate transcription.
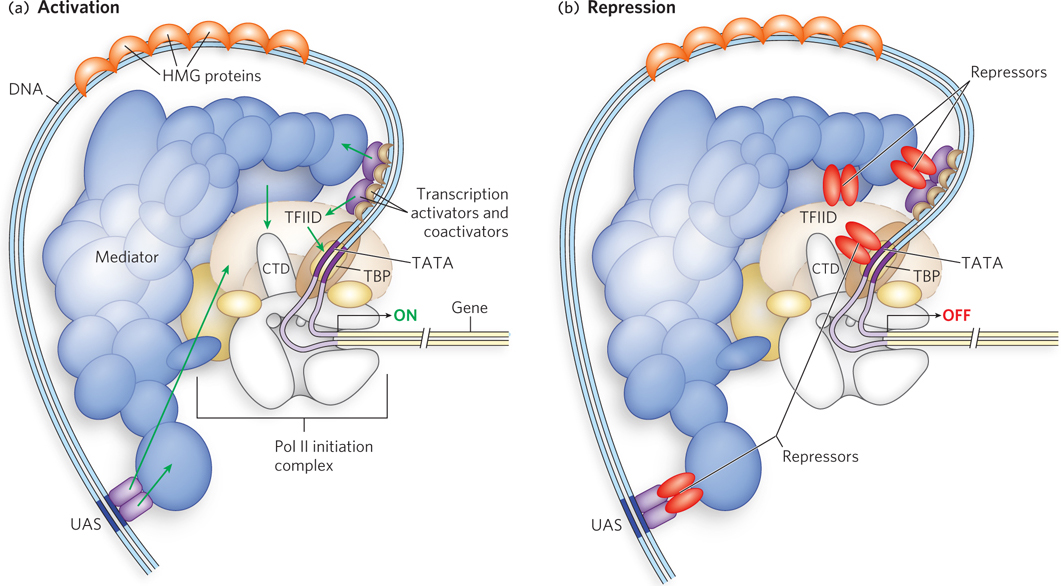
Figure 21-5: Mechanisms of activation and repression of eukaryotic gene expression. (a) Transcription activators and coactivators bound to distant regulatory sites (enhancers and UASs) recruit components of the Pol II general (basal) transcription machinery to the promoter. Coactivators such as Mediator and TFIID are required at essentially all promoters. They function as a bridge between activators and the polymerase, and do not interact with DNA directly. (b) Repression is mediated by proteins that disrupt or prevent essential contacts between Pol II and activators or coactivators. See Figure 21-7 for greater detail.
For transcription to begin, the Pol II holoenzyme must be recruited to the promoter to form a preinitiation complex with the general transcription factors. Assembly of a preinitiation complex at a typical Pol II promoter begins with the binding of TATA-binding protein (TBP) to the TATA box. TBP, which is part of the larger transcription factor complex called TFIID, then recruits additional general transcription factors and Pol II (see Figure 15-24). The minimal preinitiation complex, however, is often insufficient for the initiation of transcription, and it generally does not form at all if the promoter is buried in chromatin. Positive regulation by transcription activators and coactivators is required. We now know that the basal Pol II machinery is not as uniform as originally thought; the individual components can vary with cell type. A well-documented example is muscle cells (see the How We Know section at the end of this chapter). Thus, different combinations of general transcription factors form a complex at a promoter and are acted on by specific activator and coactivator binding proteins, adding a further level of control to the regulation of that gene.
As noted above, binding sites for transcription activators are often located far from the promoters they regulate. Recall from Chapter 19 that the intervening DNA is looped so that the various protein complexes can interact, directly or indirectly. DNA looping is promoted by certain nonhistone proteins that are abundant in chromatin and bind nonspecifically to DNA. These high-mobility group (HMG) proteins play an important structural role in chromatin remodeling and transcriptional activation (“high mobility” refers to the proteins’ rapid electrophoretic mobility in polyacrylamide gels). A structure formed by an HMG-box domain in HMG proteins can bind directly to nucleosomes, leading to altered local chromatin structure. Figure 21-6 shows the high degree to which DNA is bent by the HMG-box DNA-binding domain of the protein HMG-D of Drosophila melanogaster, one of many DNA-interactive protein structures determined in the laboratory of Mair Churchill.
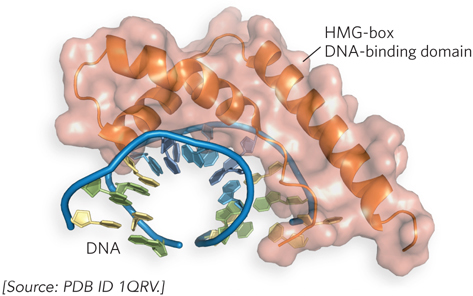
Figure 21-6: DNA looping facilitated by HMG proteins. HMG proteins bend DNA, helping form loops between enhancer and promoter elements. Binding is nonspecific. Shown here is the HMG-box DNA-binding domain of the protein HMG-D of Drosophila, bound to DNA.
In addition to transcription activators, most transcription requires coactivator protein complexes. Some major regulatory protein complexes that interact with Pol II have been defined both genetically and biochemically. They act as intermediaries between the DNA-binding transactivators and the Pol II complex (Pol II and the general transcription factors). The best-characterized coactivator is TFIID (see Chapter 15). In eukaryotes, TFIID includes TBP and 10 or more TBP-associated factors (TAFs). Some TAFs resemble histones and may play a role in competing with and thus displacing nucleosomes during the activation of transcription. Many DNA-binding transactivators aid in transcription initiation by interacting with one or more TAFs.
Another important coactivator is the Mediator complex (see Figure 15-25), which consists of 20 core polypeptides that are highly conserved from fungi to humans. Mediator binds tightly to the C-terminal domain (CTD) of the largest Pol II subunit. The Mediator complex is required for both basal and regulated transcription at Pol II promoters, and it also stimulates phosphorylation of the Pol II CTD by the general transcription factor TFIIH. Phosphorylation of the CTD enhances the efficiency of Pol II. As with TFIID, some DNA-binding transactivators interact with one or more components of the Mediator complex. Some promoters require both Mediator and TFIID coactivators. The coactivator complexes function at or near the promoter’s TATA box.
We can now begin to piece together the sequence of transcriptional activation events at a typical Pol II promoter. Crucial remodeling of the chromatin takes place in stages. Some DNA-binding transactivators have significant affinity for their binding sites even when the sites are within condensed chromatin. Binding of one transactivator may facilitate the binding of others, gradually displacing some of the nucleosomes that previously obscured the relevant DNA.
The bound transcription activators may have HAT activity or may recruit HATs or enzyme complexes such as SWI/SNF, accelerating the remodeling of surrounding chromatin (Figure 21-7). In this way, transcription activator binding can lead to the stepwise assembly of components necessary for further chromatin remodeling, to permit the transcription of specific genes. The bound transactivators, acting through complexes such as TFIID or Mediator (or both), stabilize the binding of Pol II and its associated general transcription factors, greatly facilitating formation of the preinitiation complex. Complexity in these regulatory circuits is the rule rather than the exception, with multiple DNA-bound transactivators promoting transcription.
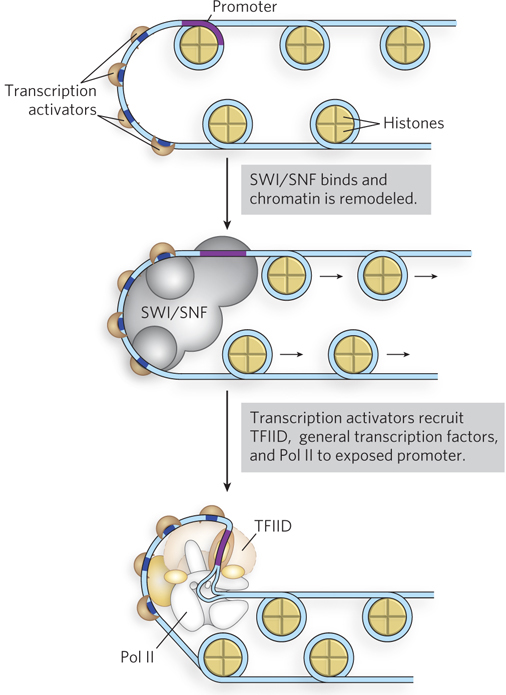
Figure 21-7: Transcription activator–mediated chromatin remodeling. Transcription activators can remodel chromatin structure by mobilizing nucleosomes; nucleosome repositioning is influenced by histone modifications. Some transcription activators have HAT activity or recruit enzyme complexes such as SWI/SNF, accelerating the remodeling of chromatin by relocating nucleosomes near a promoter. This leads to recruitment of the transcription machinery to newly exposed promoters, stimulating transcription.
The script can change from one promoter to another, but most promoters seem to require a precisely ordered assembling of components to initiate transcription. The assembly process is not always fast: at some genes it may take minutes, but it can take days at certain genes in higher eukaryotes.
Although rarer, some eukaryotic regulatory proteins that bind Pol II promoters can act as repressors, inhibiting the formation of active preinitiation complexes. Some transcription activators can adopt different conformations, enabling them to serve as activators or repressors. For example, some steroid hormone receptors function in the nucleus as DNA-binding transactivators, stimulating transcription of certain genes when a particular steroid hormone signal is present (see Section 21.3). When the hormone is absent, the receptor proteins revert to a repressor conformation, preventing formation of preinitiation complexes. In some cases this repression involves interaction with HDACs and other proteins that help restore the surrounding chromatin to its transcriptionally inactive state.
SECTION 21.1 SUMMARY
Most eukaryotic genes are inactive in their ground state, as histones cover the DNA, and are under positive control; they require multiple activator proteins to stimulate transcription.
The eukaryotic RNA polymerases require activator binding to promoter sequences to activate gene expression. The cell produces only the activator proteins necessary for transcription of the subset of genes needed at that time.
Many Pol II promoters include the TATA box and Inr sequences, as well as other sequences located far from the promoter. When bound by the appropriate regulatory proteins, these distant regulatory sequences—enhancers in higher eukaryotes and upstream activator sequences in yeast—function at the promoter through DNA looping, increasing transcription regardless of their orientation in the DNA. The DNA bending is facilitated by HMG proteins.
Transcription is stimulated by interactions between RNA polymerase core subunits and transcription activators (transactivators) bound to enhancer sequences. Often, coactivator complexes such as TFIID or Mediator act as bridges between the core transcription machinery and transactivators.







