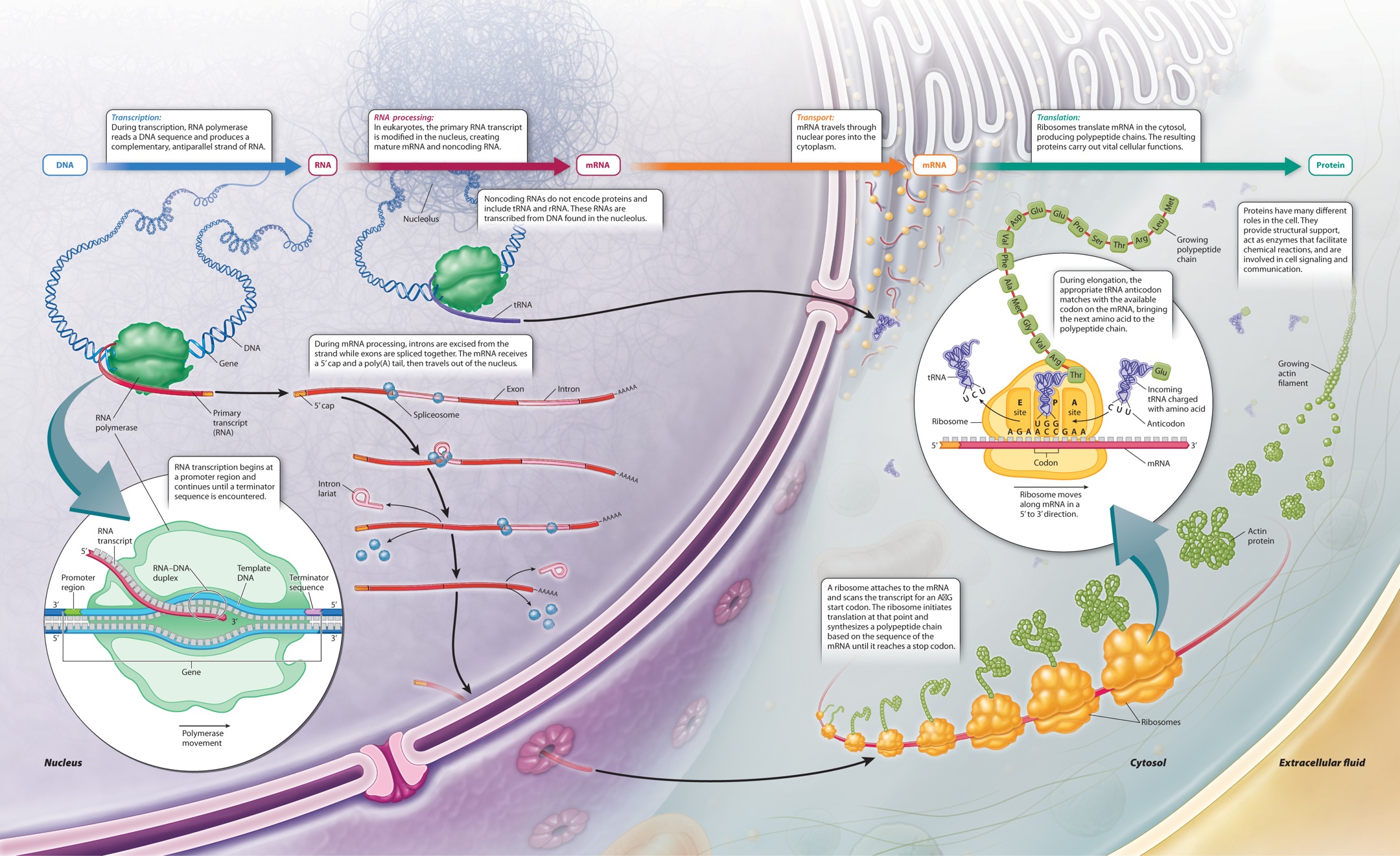
4.2 TRANSLATION: HOW PROTEINS ARE SYNTHESIZED
The three-dimensional structure of a protein determines what it can do and how it works, and the immense diversity in the tertiary and quaternary structures among proteins explains their wide range of functions in cellular processes. Yet it is the sequence of amino acids along a polypeptide chain—its primary structure—that governs how the molecule folds into a stable three-dimensional configuration. How is the sequence of amino acids specified? It is specified by the sequence of nucleotides in the DNA, in coded form. The decoding of the information takes place according to the central dogma of molecular biology, which defines information flow in a cell from DNA to RNA to protein (Figure 4.11). The key steps are known as transcription and translation. In transcription, the sequence of bases along part of a DNA strand is used as a template in the synthesis of a complementary sequence of bases in a molecule of RNA, as described in Chapter 3. In translation, the sequence of bases in an RNA molecule known as messenger RNA (mRNA) is used to specify the order in which successive amino acids are added to a newly synthesized polypeptide chain.
4.2.1 Translation uses many molecules found in all cells.
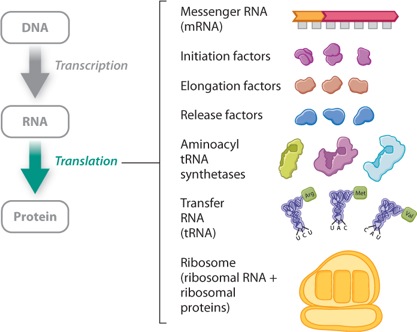
Translation requires many components. Well over 100 genes encode components needed for translation, some of which are shown in Figure 4.11. What are these needed components? First, the cell needs ribosomes, which are complex structures of RNA and protein. Ribosomes bind with mRNA, and it is on ribosomes that translation takes place. In prokaryotes, translation occurs as soon as the mRNA comes off the DNA template. In eukaryotes, the processes of transcription and translation are physically separated: Transcription takes place in the nucleus, and translation takes place on ribosomes in the cytoplasm.
4-9
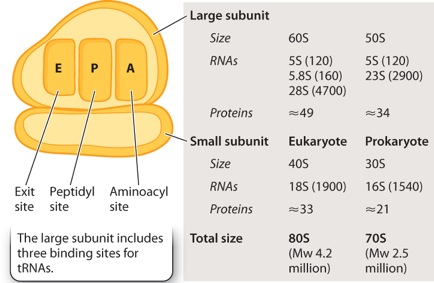
In both eukaryotes and prokaryotes, the ribosome consists of a small subunit and a large subunit, each composed of both RNA and protein. The sizes of the subunits are given in Svedberg units (S), a measure of size and shape. Eukaryotic ribosomes are larger than prokaryotic ribosomes. As indicated in Figure 4.12, the large subunit of the ribosome includes three binding sites for molecules of transfer RNA (tRNA), which are called the A (aminoacyl) site, the P (peptidyl) site, and the E (exit) site.
A major role of the ribosome is to ensure that, when the mRNA is in place on the ribosome, the sequence in the mRNA coding for amino acids is read in successive, non-overlapping groups of three nucleotides, much as you would read the sentence
THEBIGBOYSAWTHEBADMANRUN
Each non-overlapping group of three adjacent nucleotides (like THE or BIG or BOY, for example) constitutes a codon, and each codon in the mRNA codes for a single amino acid in the polypeptide chain.
In the example above, it is clear that the sentence begins with THE. However, in a long linear mRNA molecule, the ribosome could begin at any of three possible positions. These are known as reading frames. As an analogy, if the letters THE were the start codon for reading text, then we would know immediately how to read
ZWTHEBIGBOYSAWTHEBADMANRUN
However, without knowing the first codon of this string of letters, we could find three ways to break the sentence into three-letter words:

Obviously, only one of these frames is correct. The same is true for mRNAs.
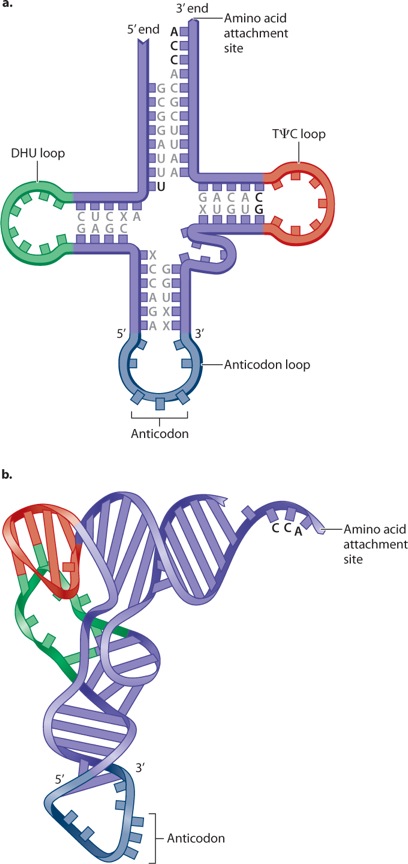
While the ribosome establishes the correct reading frame for the codons, the actual translation of each codon in the mRNA into one amino acid in the polypeptide is carried out by means of transfer RNA (tRNA). Transfer RNAs are small RNA molecules of 70 to 90 nucleotides (Figure 4.13). Each has a characteristic self-pairing structure that can be drawn as a cloverleaf, as shown in Figure 4.13a, but the actual structure is more like that in Figure 4.13b. In Figure 4.13a, the bases found in all tRNA molecules are indicated by the letters; the Greek letter ψ (psi) stands for pseudouracil, a slightly modified form of uracil. Three bases in the anticodon loop make up the anticodon; these are the three nucleotides that undergo base pairing with the corresponding codon.
4-10
Each tRNA has the nucleotide sequence CCA at its 3′ end (Figure 4.13a), and the 3′ hydroxyl of the A is the attachment site for the amino acid corresponding to the anticodon. Enzymes called aminoacyl tRNA synthetases connect specific amino acids to specific tRNA molecules (Figure 4.14). Therefore, they are directly responsible for actually translating the codon sequence in a nucleic acid to a specific amino acid in a polypeptide chain. Most organisms have one aminoacyl tRNA synthetase for each amino acid. The enzyme binds to multiple sites on any tRNA that has an anticodon corresponding to the amino acid, and it catalyzes formation of the covalent bond between the amino acid and tRNA. A tRNA that has no amino acid attached is said to be uncharged, and one with its amino acid attached is said to be charged. Amino acid tRNA synthetases are very accurate and attach the wrong amino acid far less often than 1 time in 10,000. This accuracy is greater than that of subsequent steps in translation.
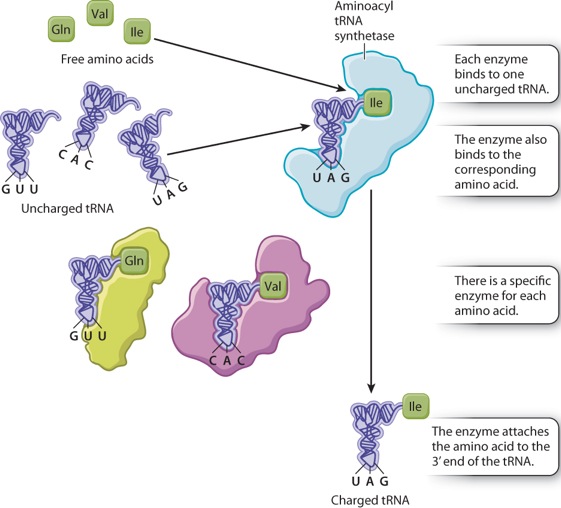
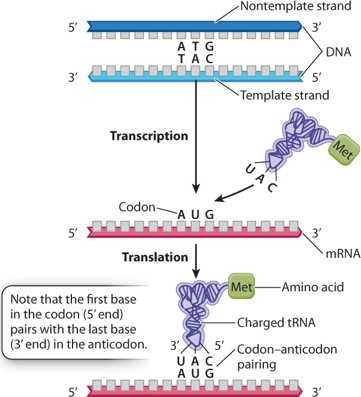
Although the specificity for attaching an amino acid to the correct tRNA is a property of aminoacyl tRNA synthetase, the specificity of DNA–RNA and codon–anticodon interactions result from base pairing. Figure 4.15 shows the relationships for one codon in double-stranded DNA, in the corresponding mRNA, and in the codon–anticodon pairing between the mRNA and the tRNA. Note that the first (5′) base in the codon in mRNA pairs with the last (3′) base in the anticodon because, as noted in Chapter 3, nucleic acid strands that undergo base pairing must be antiparallel.
4.2.2 The genetic code shows the correspondence between codons and amino acids.
Figure 4.15 shows how the codon AUG specifies the amino acid methionine (Met) by base pairing with the anticodon of a charged tRNA, denoted tRNAMet. Most codons specify an amino acid according to a genetic code. This code is sometimes called the “standard” genetic code because, while it is used by almost all cells, some minor differences are found in a few organisms as well as in mitochondria.
4-11
The codon that initiates the process of translation is AUG, which specifies Met. The polypeptide is synthesized from the amino end to the carboxyl end, and so Met forms the amino end of any polypeptide being synthesized; however, in many cases the Met is cleaved off by an enzyme after synthesis is complete. The AUG codon is also used to specify the incorporation of Met at internal sites within the polypeptide chain.
As is apparent in Figure 4.15, the AUG codon that initiated translation is preceded by a region in the mRNA that is not translated. The position of the initiator AUG codon in the mRNA establishes the reading frame that determines how the downstream codons (those following the AUG) are to be read.
Once the initial Met creates the amino end of a new polypeptide chain, the downstream codons are read one by one in non-overlapping groups of three bases. At each step, the ribosome binds to a tRNA with an anticodon that can base pair with the codon, and the amino acid on that tRNA is attached to the growing chain to become the new carboxyl end of the polypeptide chain. This process continues until one of three “stop” codons is encountered: UAA, UAG, or UGA. (The stop codons are also called termination codons or sometimes nonsense codons.) At this point, the polypeptide is finished and released into the cytosol.
The standard genetic code was deciphered in the 1960s by a combination of techniques, but among the most ingenious was the development by Har Gobind Khorana and his colleagues of chemical methods for making synthetic RNAs of known sequence. This experiment is illustrated in Figure 4.16.
Figure 4.16: How was the genetic code deciphered?
METHOD Har Gobind Khorana and his group made RNAs of known sequence. They then added these synthetic RNAs to a solution containing all of the other components needed for translation. By adjusting the concentration of magnesium and other factors, the researchers could get the ribosome to initiate synthesis with any codon, even if not AUG.
EXPERIMENT 1 AND RESULTS When a synthetic poly(U) was used as the mRNA, the resulting polypeptide was polyphenylalanine (Phe–Phe–Phe…):
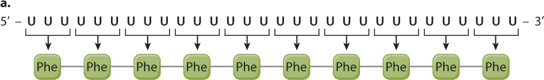
CONCLUSION The codon UUU corresponds to Phe. The poly(U) mRNA can be translated in three possible reading frames, depending on which U is the 5′ end of the start codon, but in each of them, all the codons are UUU.
EXPERIMENT 2 AND RESULTS When a synthetic mRNA with alternating U and C was used, the resulting polypeptide had alternating serine (Ser) and leucine (Leu):
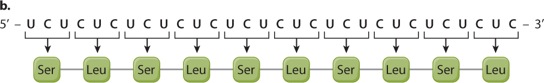
CONCLUSION Here again there are three reading frames, but each of them has alternating UCU and CUC codons. The researchers could not deduce from this result whether UCU corresponds to Ser and CUC to Leu or the other way around; the correct assignment came from experiments using other synthetic mRNA molecules.
EXPERIMENT 3 AND RESULTS When a synthetic mRNA with repeating UCA was used, three different polypeptides were produced—polyserine (Ser), polyhistidine (His), and polyisoleucine (Ile).
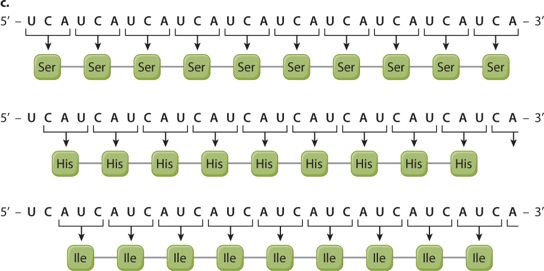
CONCLUSION The results do not reveal which of the three reading frames corresponds to which amino acid, but this was sorted out by studies of other synthetic polymers.
SOURCE Khorana, H. G. 1972. “Nucleic Acid Synthesis in the Study of the Genetic Code.” In Nobel Lectures, Physiology or Medicine 1963–1970. Amsterdam: Elsevier, 1972.
4-12
→ Quick Check 2 What polypeptide sequences would you expect to result from a synthetic mRNA with the repeating sequence 5′-UUUGGGUUUGGGUUUGGG-3″?
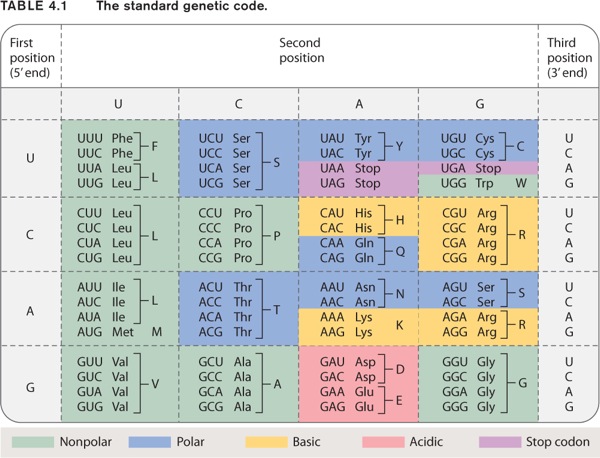
The standard genetic code shown in Table 4.1 has 20 amino acids specified by 64 codons. Many amino acids are therefore specified by more than one codon, and hence the genetic code is redundant, or “degenerate.” The redundancy has strong patterns, however:
- The redundancy results almost exclusively from the third codon position.
- When an amino acid is specified by two codons, they differ either in whether the third position is a U or a C (both pyrimidine bases), or in whether the third position is an A or a G (both purine bases).
- When an amino acid is specified by four codons, the identity of the third codon position does not matter; it could be U, C, A, or G.
The chemical basis of these patterns results from two features of translation. First, in many tRNA anticodons the 5′ base that pairs with the 3′ (third) base in the codon is chemically modified into a form that can pair with two or more bases in the codon. Second, in the ribosome, there is less-than-perfect alignment between the third position of the codon and the base that pairs with it in the anticodon, so the requirements for base pairing are somewhat relaxed; this feature of the codon–anticodon interaction is referred to as wobble.
4.2.3 Translation consists of initiation, elongation, and termination.
Translation is usually divided into three separate processes. The first is initiation, in which the initiator AUG codon is recognized and Met is established as the first amino acid in the new polypeptide chain. The second process is elongation, in which successive amino acids are added one by one to the growing chain. And the third process is termination, when the addition of amino acids stops and the completed polypeptide chain is released from the ribosome.
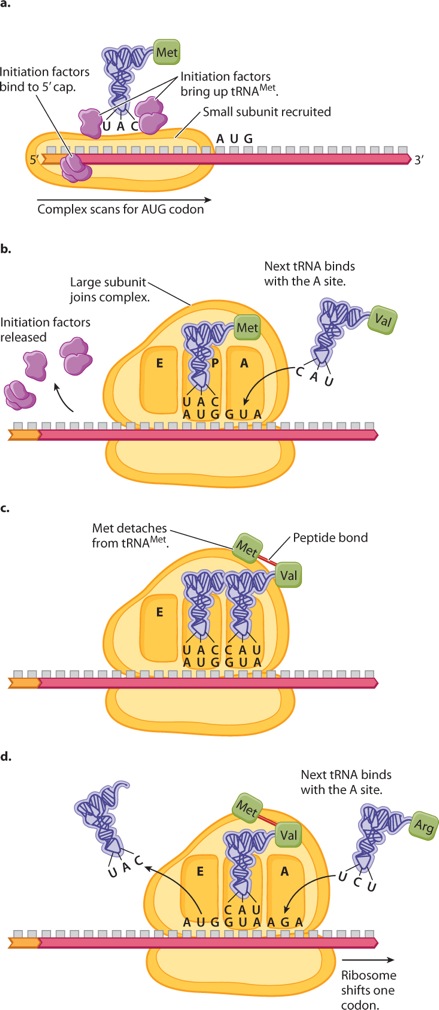
Initiation of translation (Figure 4.17) requires a number of protein initiation factors that bind to the mRNA. In eukaryotes, one group of initiation factors binds to the 5′ cap that is added to the mRNA during processing (Figure 4.17a). These recruit a small subunit of the ribosome, and other initiation factors bring up a transfer RNA charged with methionine (Met). The initiation complex then moves along the mRNA until it encounters the first AUG triplet. The position of this AUG establishes the translational reading frame.
When it is encountered, a large ribosomal subunit joins the complex, the initiation factors are released, and the next tRNA is ready to join the ribosome (Figure 4.17b). Note in Figure 4.17b that the tRNAMet binds with the P (peptidyl) site in the ribosome and that the next tRNA in line comes in at the A (aminoacyl) site. Once the new tRNA is in place, a coupled reaction takes place in which the bond connecting the Met to its tRNA is transferred to the amino group of the next amino acid in line as the first peptide bond is formed (Figure 4.17c). The new peptide is now attached to the tRNA in the A site. Formation of the peptide bond requires multiple proteins in the large subunit, but an RNA in the large subunit is the actual catalyst. The ribosome then shifts one codon to the right (Figure 4.17d), which moves the uncharged tRNAMet to the E site and the peptide-bearing tRNA to the P site, freeing the A site for the next charged tRNA in line to come in.
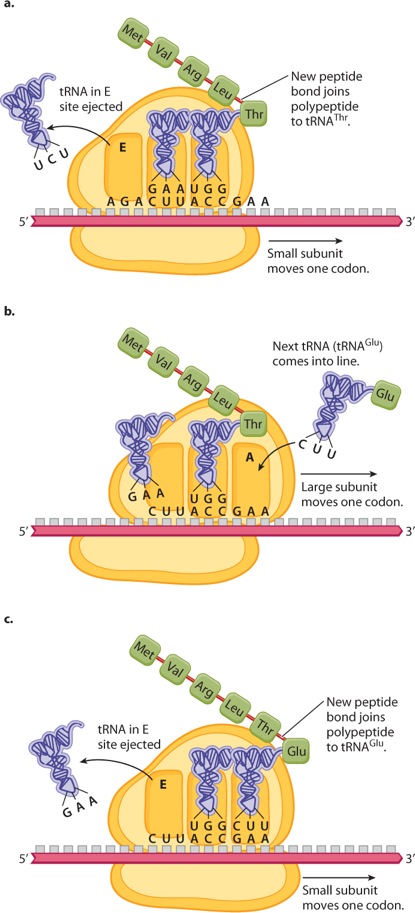
Once translation has been initiated, it proceeds step by step through the process of elongation (Figure 4.18). Figure 4.18a shows the situation immediately after the formation of a new peptide bond that attaches the growing polypeptide chain to the amino acid linked to the tRNA in the A site (in this case tRNAThr); the small ribosomal subunit has moved one codon farther along the mRNA, and the tRNA in the E (exit) site is being ejected. Figure 4.18b shows the situation an instant later, when the large ribosomal subunit has moved downstream one codon, emptying the A site by shifting the tRNAThr to the P site in the middle and the tRNALeu to the E site. Note that, while there are three tRNA binding sites on the ribosome, only two of them are occupied at any one time. Figure 4.18c shows the situation slightly later; it is almost identical to that in Figure 4.18a, except that the polypeptide is one amino acid longer and the ribosome has moved one codon farther to the right. Ribosome movement along the mRNA and formation of the peptide bonds require energy, which is obtained by breaking the high-energy bonds of the molecule GTP bound with proteins called elongation factors.
4-13
4-14
The elongation process shown in Figure 4.18 continues until one of the stop codons (UAA, UAG, or UGA) is encountered; these codons signal termination of polypeptide synthesis. Termination takes place because the stop codons do not have corresponding tRNA molecules. Rather, when a stop codon is encountered, a protein release factor binds to the A site of the ribosome. The release factor causes the bond connecting the polypeptide to the tRNA to break, which creates the carboxyl terminus of the polypeptide and completes the chain. Once the finished polypeptide is released, the small and large ribosomal subunits disassociate from the mRNA and from each other.
Although elongation and termination are very similar in prokaryotes and in eukaryotes, translation initiation differs between the two (Figure 4.19). In eukaryotes, the initiation complex forms at the 5′ cap and scans along the mRNA until the first AUG is encountered (Figure 4.19a). In prokaryotes, the mRNA molecules have no 5′ cap. Instead, the initiation complex is formed at one or more internal sequences present in the mRNA known as a Shine–Dalgarno sequence (Figure 4.19b). In E. coli, the Shine–Dalgarno sequence is 5′-AGGAGGU-3′, and it is followed by an AUG codon eight nucleotides farther downstream that serves as an initiation codon for translation. The ability to initiate translation internally allows prokaryotic mRNAs to contain open reading frames for more than one protein. Such an mRNA is known as a polycistronic mRNA. In Figure 4.19b, the polycistronic mRNA codes for three different polypeptide chains, each with its own AUG initiation codon preceded eight nucleotides upstream by its own Shine–Dalgarno sequence. Each Shine–Dalgarno sequence can serve as an initiation site for translation, and so all three polypeptides can be translated.
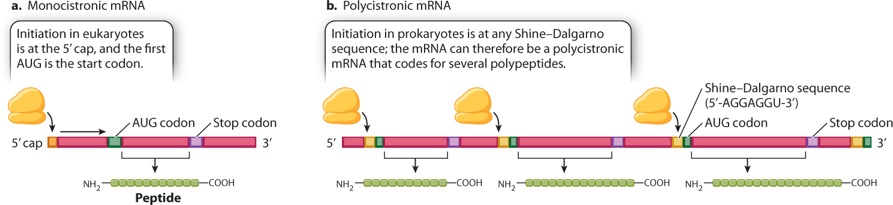
A polycistronic mRNA results from transcription of a group of functionally related genes located in tandem along the DNA and transcribed as a single unit from one promoter; this type of gene organization is known as an operon. Prokaryotes have many of their genes organized into operons because the production of a polycistronic mRNA allows all the protein products to be expressed together whenever they are needed. Typically, the genes organized into operons are those whose products are needed either for successive steps in the synthesis of an essential small molecule, such as an amino acid, or else for successive steps in the breakdown of a source of energy, such as a complex carbohydrate.
→ Quick Check 3 Bacterial DNA containing an operon encoding three enzymes is introduced into chromosomal DNA in yeast (a eukaryote) in such a way that it is properly flanked by a promoter and a transcriptional terminator. The bacterial DNA is transcribed and the RNA correctly processed, but only the protein nearest the promoter is produced. Can you suggest why?
4.2.4 How did the genetic code originate?

During transcription and translation, proteins and nucleic acids work together to convert the information stored in DNA into proteins. If we think about how such a system might have originated, however, we immediately confront a chicken-and-egg problem: Cells need nucleic acids to make proteins, but proteins are required to make nucleic acids. Which came first? In Chapter 2, we discussed the special features that make RNA an attractive candidate for both information storage and catalysis in early life. Early in evolutionary history, then, proteins had to be added to the mix. No one fully understands how they were incorporated, but researchers are looking closely at tRNA, the molecule that completes the “translating” step of translation.
4-15
In modern cells, tRNA shuttles amino acids to the ribosome, but an innovative hypothesis suggests that in early life tRNA-like molecules might have served a different function. This proposal holds that the early precursors of the ribosome were RNA molecules that facilitated the replication of other RNAs, not proteins. In this version of an RNA world, precursors to tRNA would have shuttled nucleotides to growing RNA strands. Researchers hypothesize that tRNAs bound to amino acids may have acted as simple catalysts, facilitating more accurate RNA synthesis. Through time, amino acids brought into close proximity in the process of building RNA molecules might have polymerized to form peptide chains. From there, natural selection would favor the formation of peptides that enhanced replication of RNA molecules, bringing proteins into the chemistry of life.
All of the steps in gene expression, including transcription and translation, are summarized in Figure 4.20, in the next section.
Visual Summary: Gene Expression