Concept 11.1: Many Prokaryotic Genes Are Regulated in Operons
In Chapter 10 we introduced the concepts of gene expression. DNA is initially expressed as RNA, and in many cases the RNA is then translated into protein by the ribosome. Throughout this book we describe instances where gene expression is altered so that the level of protein produced from a particular gene is altered. Such changes are influenced by environmental conditions and the developmental stage of the cell or organism. Here are a few examples:
- In Chapter 5: When an extracellular signal binds to its receptor on a eukaryotic cell, it sets in motion a signal transduction pathway that may end with some genes being activated (their expression switched on) or others being repressed (their expression switched off).
- In Chapter 7: During the cell cycle, cyclins are synthesized only at specific stages. The genes for cyclins are inactive at other stages in the cycle.
- In Chapter 9: When a virus infects a host cell, it can “hijack” the host gene expression machinery and divert it to viral gene expression.
These and other examples indicate that gene expression is precisely regulated.
Genes are subject to positive and negative regulation
At every step of the way from DNA to protein, gene expression can be regulated (FIGURE 11.1). As we proceed through this chapter, you will see examples of gene regulation at the transcriptional, posttranscriptional, translational, and post-translational levels. An important form of gene regulation is at the level of transcription.
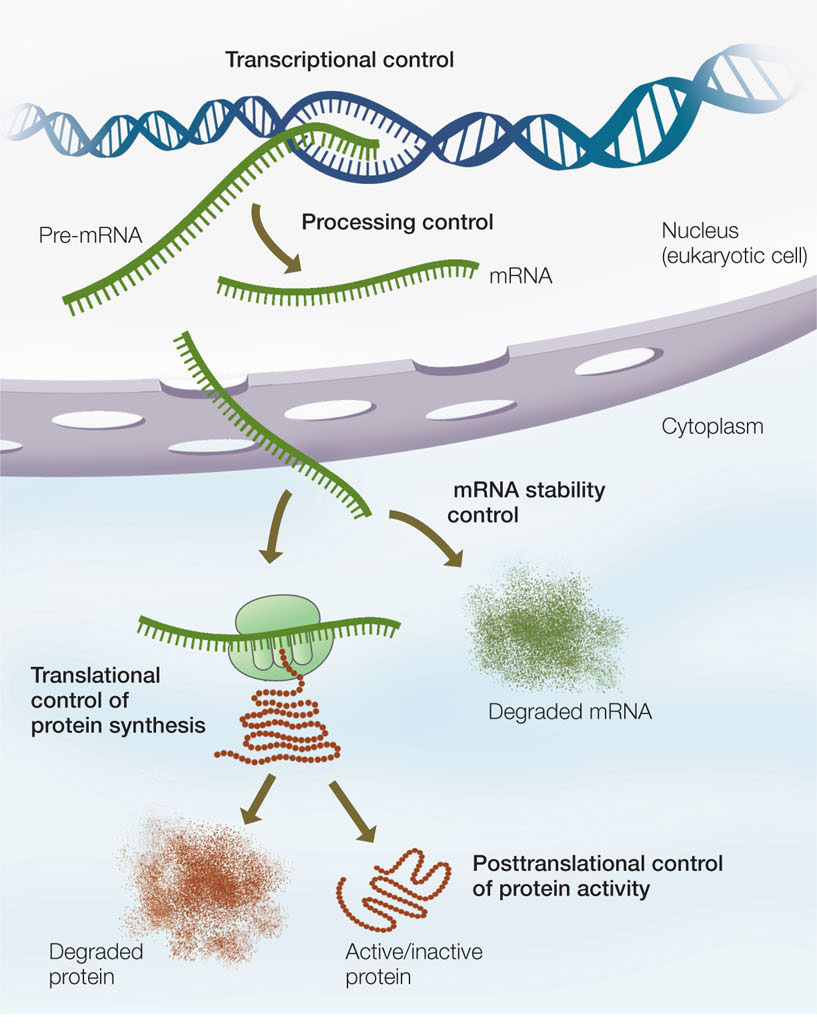
Go to ACTIVITY 11.1 Eukaryotic Gene Expression Control Points
PoL2e.com/ac11.1
Gene expression begins at the promoter, a region of DNA containing the site where RNA polymerase binds to initiate transcription. As we mentioned above, not all genes are active (being transcribed) at a given time. Two types of regulatory proteins—called transcription factors—control whether or not a gene is active: repressors and activators. These proteins bind to specific DNA sequences at or near the promoter (FIGURE 11.2):
- In negative regulation, a repressor binds a specific site in or near the promoter to prevent transcription.
- In positive regulation, the binding of an activator stimulates transcription.
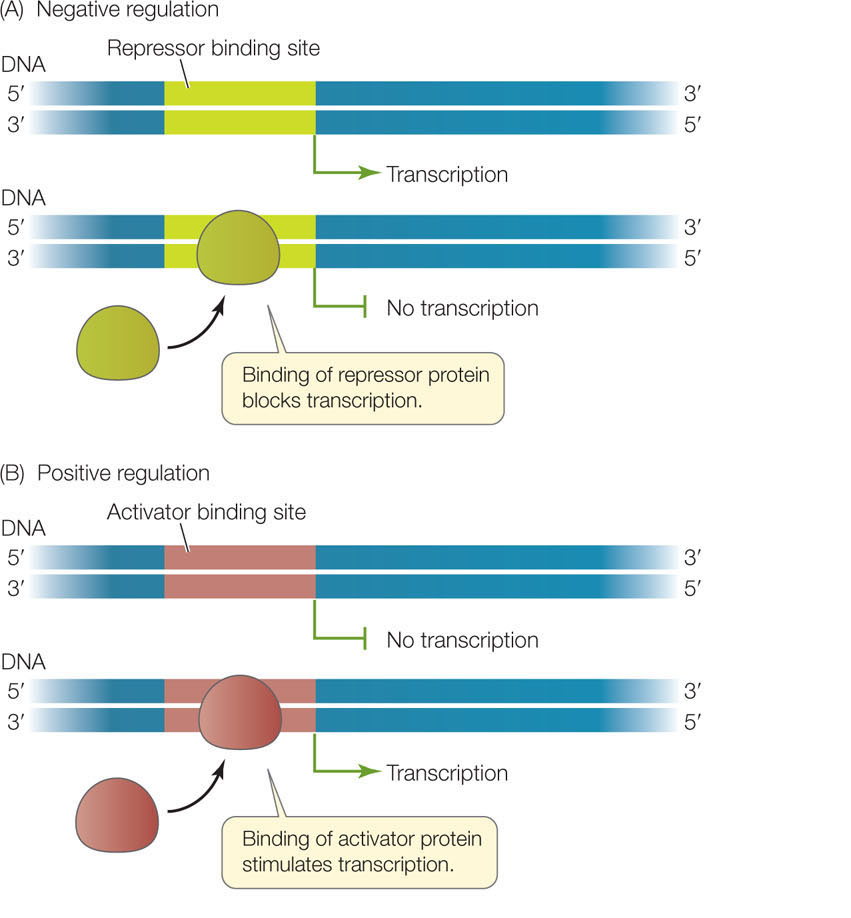
You will see these mechanisms, or combinations of them, as we examine the regulation of prokaryotic, eukaryotic, and viral genes. Let’s begin by looking at the regulation of gene expression in prokaryotes.
LINK
You can review the processes of transcription in Concept 10.2
Regulating gene transcription is a system that conserves energy
Prokaryotes conserve energy and resources by making certain proteins only when they are needed. Because their environments can change abruptly, prokaryotes have evolved mechanisms to rapidly alter the expression levels of certain genes when conditions warrant. An example is the bacterium Escherichia coli, which normally inhabits the intestines of humans and other mammals. E. coli must be able to adjust to sudden changes in its chemical environment as the foods consumed by its host change (for example, from a meal containing glucose at one time to one containing lactose at another). In many cases E. coli responds to such changes by changing the transcription of its genes. To illustrate this we will look at the regulation of the pathway for lactose catabolism in E. coli.
217
Lactose is a β-galactoside—a disaccharide containing galactose linked to glucose. Three proteins are involved in the initial uptake and metabolism of lactose by E. coli:
- β-galactoside permease is a carrier protein in the bacterial cell membrane that moves the sugar into the cell.
- β-galactosidase is an enzyme that hydrolyzes lactose to glucose and galactose.
- β-galactoside transacetylase transfers acetyl groups from acetyl CoA to certain β-galactosides. Its role in the metabolism of lactose is not clear.
When E. coli is grown on a medium that contains glucose but no lactose, the basal (uninduced) levels of these three proteins are extremely low—only a few molecules per cell. But if the cells are transferred to a medium with lactose as the predominant sugar, they promptly begin making all three proteins after a short lag period, and within 10 minutes there are about 3,000 of each of these proteins per cell (the induced level):
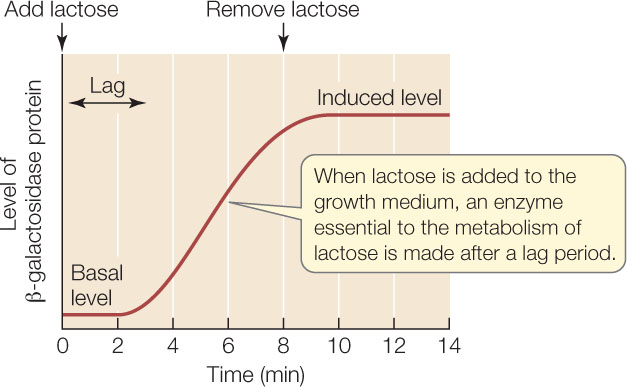
What causes this dramatic increase? A clue comes from measuring the concentration of mRNA for β-galactosidase. After lactose is added to the medium, the mRNA level increases before the level of β-galactosidase protein begins to rise:
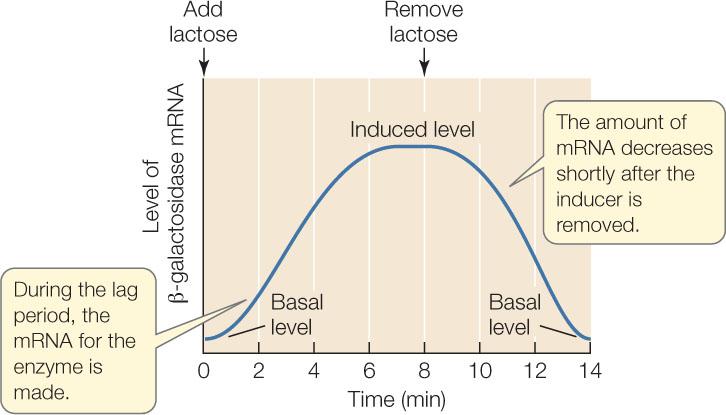
The mRNA is produced during the lag phase and then is translated into protein. The high mRNA level depends on the presence of lactose, because if lactose is removed, the mRNA level goes down. The response of the bacteria to lactose is clearly at the level of transcription. (The level of β-galactosidase protein does not go down immediately after the inducer is removed because the protein is more stable than the mRNA.)
Compounds that stimulate the transcription of specific genes are called inducers, and genes that can be activated by inducers are called inducible genes. In contrast, some other genes are expressed most of the time at a constant rate; these are called constitutive genes. The lactose-metabolizing proteins in E. coli are encoded by inducible genes. When lactose first enters the cell, some of it is converted to a similar molecule called allolactose. Allolactose is the inducer that switches on the expression of the genes for the lactose-metabolizing proteins.
Operons are units of transcriptional regulation in prokaryotes
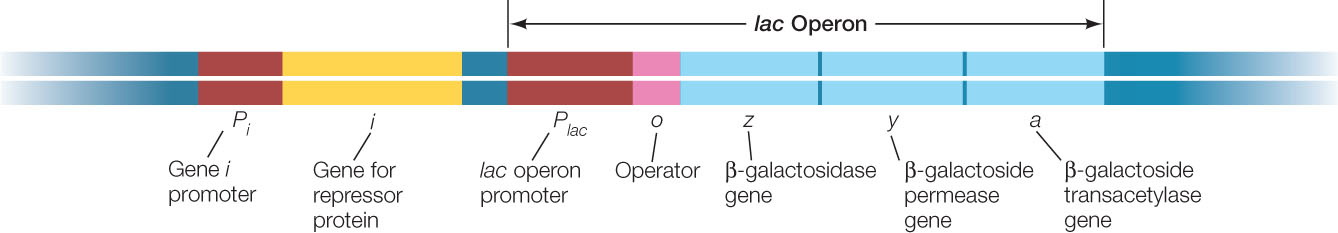
The genes that encode the three proteins for processing lactose in E. coli lie adjacent to one another on the E. coli chromosome. This arrangement—which is common for functionally related genes in prokaryotes—is no coincidence: the genes share a single promoter, and their DNA is transcribed into a single, continuous molecule of mRNA that contains the coding regions for the three proteins. Because this particular mRNA governs the synthesis of all three lactose-metabolizing enzymes, either all or none of these enzymes are made at any particular time.
A cluster of genes with a single promoter is called an operon, and the operon that encodes the three lactose-metabolizing enzymes in E. coli is called the lac operon. The lac operon promoter can be very efficient (the maximum rate of mRNA synthesis can be high), but its activity can be reduced when the enzymes are not needed. This example of transcriptional regulation, which we explore in more detail below, was worked out in the 1960s by Nobel Prize winners François Jacob and Jacques Monod.
218
Operator-repressor interactions regulate transcription in the lac and trp operons
The lac operon has a DNA sequence called an operator, which is near the promoter and controls transcription of the lac genes (FIGURE 11.3). An operator is a repressor-binding site that can bind very tightly with a repressor protein (see Figure 11.2A). Repressors play different roles in different operons:
- An inducible operon is turned off unless needed.
- A repressible operon is turned on unless not needed.
In the case of the inducible lac operon, a repressor protein prevents transcription until the lac-encoded proteins are needed. In contrast, the trp operon (described below) is a repressible operon that is turned off by a repressor only under particular circumstances.
{em}lac{/em} Operon
As we described above, the lac operon is not transcribed at high levels unless a β-galactoside such as lactose is the predominant sugar available in the cell’s environment. A repressor protein normally binds to the operator, preventing RNA polymerase from binding and thereby blocking transcription. When lactose is present, the repressor detaches from the operator, allowing RNA polymerase to bind to the promoter and start transcribing the lac genes (FIGURE 11.4).
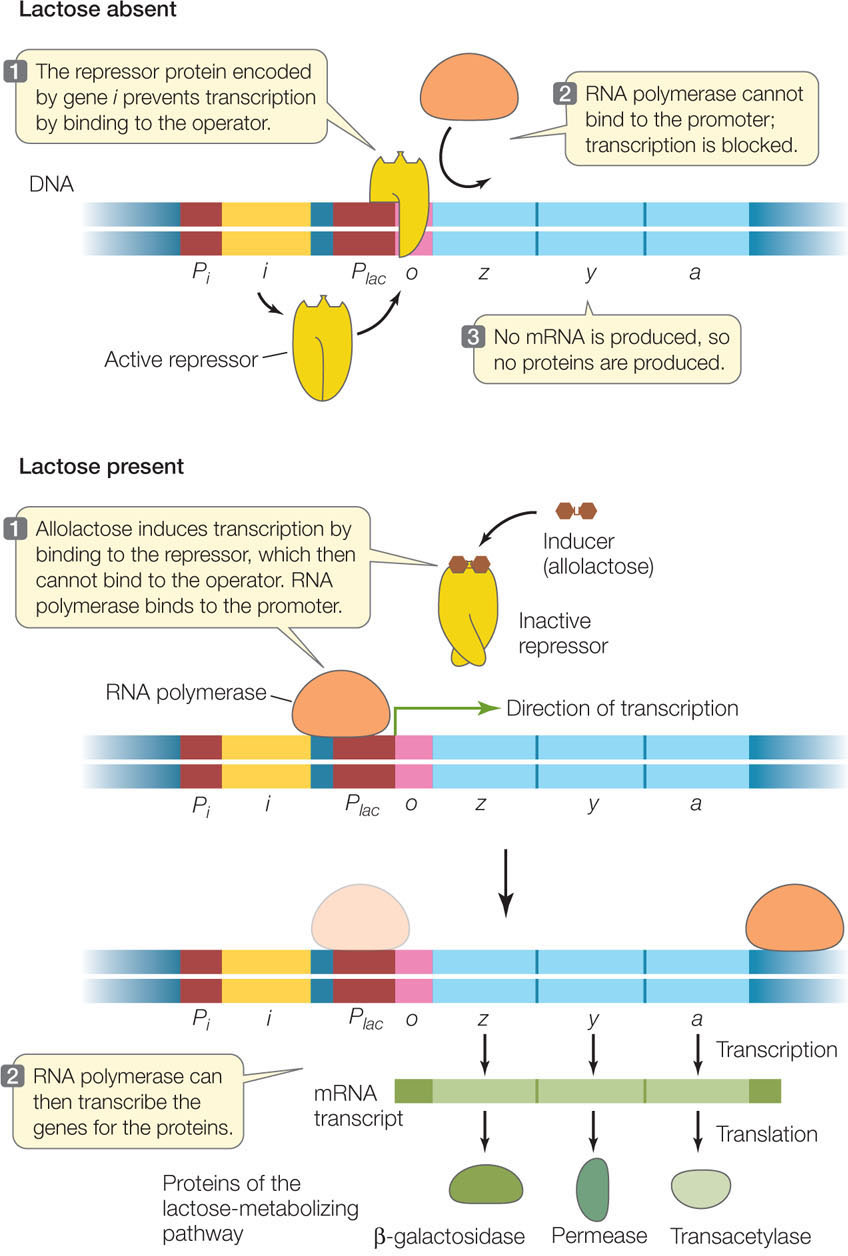
The key to this regulatory system is the repressor protein. Expressed from a constitutive gene (one that is always active), the repressor is always present in the cell in adequate amounts to occupy the operator and keep the operon turned off. The repressor has a recognition site for the DNA sequence in the operator, and it binds very tightly. However, it also has an allosteric binding site for the inducer. When the inducer (allolactose) binds to the repressor, the repressor changes shape so that it can no longer bind DNA.

Go to ANIMATED TUTORIAL 11.1 The lac Operon
PoL2e.com/at11.1
219
The gene for the lac repressor (gene i in Figure 11.3) is located upstream of the lac operon on the E. coli chromosome. The lac i gene is referred to as a regulatory gene because it encodes a regulatory protein (a transcription factor). In contrast, a structural gene is any gene that encodes a protein that is not directly involved in gene regulation. The three genes that encode the lactose-metabolizing enzymes are structural genes.
{em}trp{/em} Operon
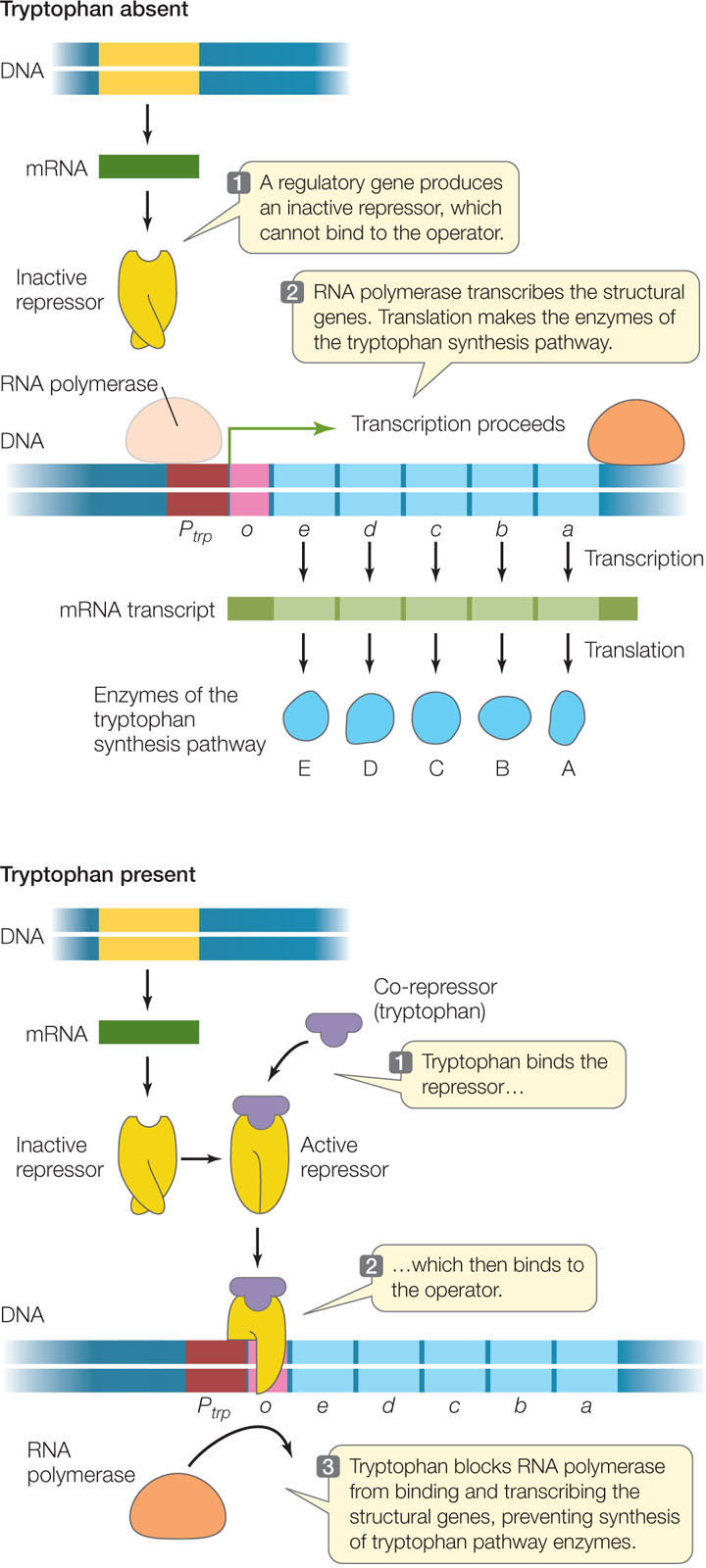
Like an inducible operon, a repressible operon is switched off when its repressor is bound to its operator. However, in this case the repressor binds to the DNA only in the presence of a corepressor. The corepressor is a molecule that binds to the repressor, causing it to change shape and bind to the operator, thereby inhibiting transcription. An example is the operon whose structural genes catalyze the synthesis of the amino acid tryptophan:
When tryptophan is present in adequate concentrations, it is energy-efficient for the cell to stop making the enzymes for tryptophan synthesis. Therefore tryptophan itself functions as a corepressor: tryptophan binds to the repressor of the trp operon, causing the repressor to bind to the trp operator to prevent transcription of the enzymes in the pathway (FIGURE 11.5).

Go to ANIMATED TUTORIAL 11.2 The trp Operon
PoL2e.com/at11.2
To summarize the differences between these two regulatory systems:
- In inducible systems, the substrate of a metabolic pathway (the inducer) interacts with a transcription factor (the repressor), rendering the repressor incapable of binding to the operator and thus allowing transcription.
- In repressible systems, a product of a metabolic pathway (the corepressor) binds to the repressor protein, which is then able to bind to the operator and block transcription.
In general, inducible systems control catabolic pathways (which are turned on only when the substrate is available), whereas repressible systems control anabolic pathways (which are turned on until the concentration of the product becomes sufficient).
LINK
You can review catabolic and anabolic reactions in Concept 2.5
In both of the systems described above, the regulatory protein is a repressor that functions by binding to the operator. Transcription in prokaryotes can also be regulated by activator proteins that bind to DNA sequences at or near the promoter and promote transcription (see Figure 11.2). Like repressors, activators can regulate both inducible and repressible systems. Furthermore, many genes and operons are controlled by more than one regulatory mechanism. We will discuss transcription factors in more detail in Concept 11.2.
We have now seen two basic systems for regulating a metabolic pathway. In Concept 3.4 we described the allosteric regulation of enzyme activity—a mechanism that allows rapid fine-tuning of metabolism. The regulation of transcription is slower but results in greater savings of energy and resources. Protein synthesis is a highly endergonic process; synthesizing mRNA, charging tRNA, and moving the ribosomes along mRNA all require large amounts of energy. FIGURE 11.6 compares allosteric and transcriptional regulation.
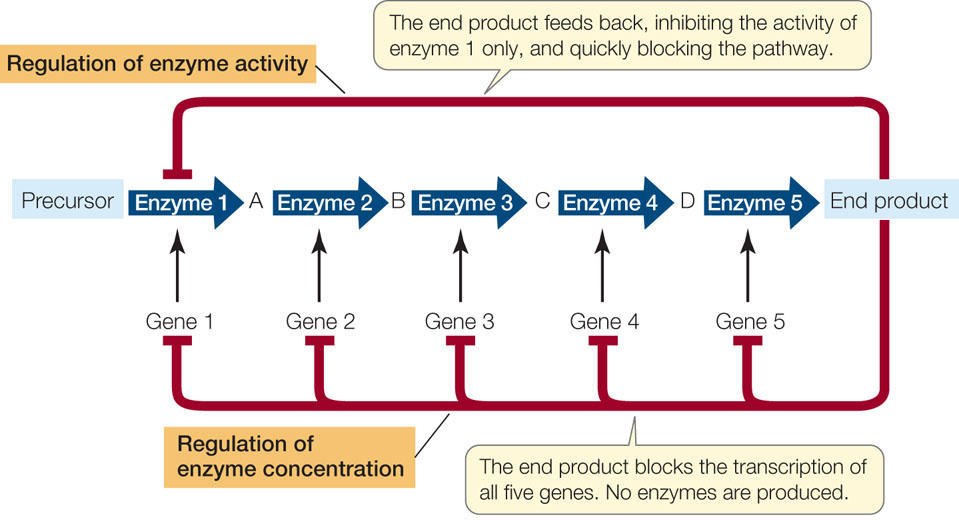
220
RNA polymerase can be directed to a class of promoters
As noted above and in Chapter 10, RNA polymerase binds to specific DNA sequences at the promoter to initiate transcription. We have just described how repressor proteins can physically block RNA polymerase binding. However, there are other proteins in prokaryotes called sigma factors that can bind to RNA polymerase and direct the polymerase to specific promoters.
Genes that encode proteins with related functions may be at different locations in the genome but have the same promoter sequence. This allows them to be expressed at the same time and under the same physiological conditions. For example, some bacteria stop growing when nutrients in their environment are depleted. When this happens, they adopt an alternative lifestyle called sporulation—they reduce their metabolic activity and form a tough spore coat (see Concept 19.2). This process involves the sequential expression of specific classes of genes. Each member of a gene class has a common promoter sequence, and RNA polymerase is directed to the promoter in each case by a specific sigma factor. As we will see in Concept 11.2, this form of global gene regulation by proteins binding to RNA polymerase is also common in eukaryotes.
Viruses use gene regulation strategies to hijack host cells
The immunologist Sir Peter Medawar once described a virus as “a piece of bad news wrapped in protein.” As we described in Concept 9.1, a virus injects its genetic material into a host cell, and in many cases it turns that cell into a virus factory:
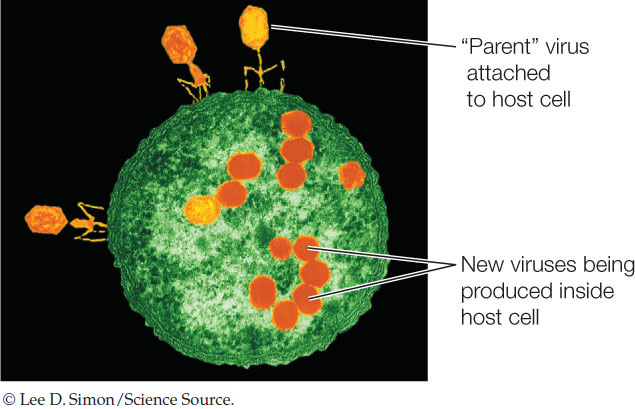
This involves a radical change in gene expression for the host cell, and can result in the death of the cell when new viral particles are released.
Viruses are not cells and do not carry out many of the processes characteristic of life. They are dependent on living cells to reproduce. Unlike living cells, not all viruses use double-stranded DNA as the genetic material that is contained within the viral particle and transmitted from one generation to the next. The viral genome may consist of double-stranded DNA, single-stranded DNA, or double- or single-stranded RNA. But whether the genetic material is DNA or RNA, the viral genome takes over the host’s protein synthetic machinery within minutes of entering the cell.
APPLY THE CONCEPT: Many prokaryotic genes are regulated in operons, which include regulatory DNA sequences
Genetic mutations are useful in analyzing the control of gene expression. In the lac operon of E. coli (see Figures 11.3 and 11.4), gene i codes for the repressor protein, Plac is the promoter, o is the operator, and z is the first structural gene. The superscript “+” designates the wild type; superscript “−” means mutant. Fill in the table, describing the level of transcription in different genetic and environmental conditions. (The first line of the table has been filled in as an example.)
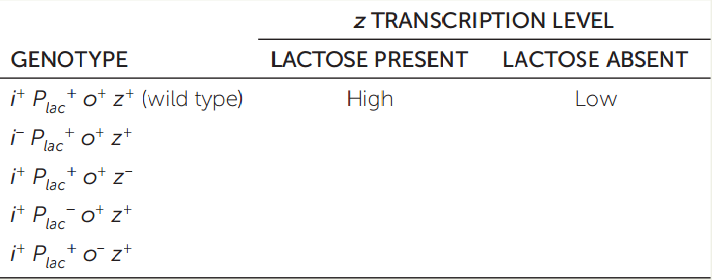
221
Typically, the host cell immediately begins to produce new viral particles (virions), which are released as the cell breaks open, or lyses. This type of prokaryotic viral life cycle is called lytic. Some viral life cycles also include a lysogenic or dormant phase. In this case the viral genome becomes incorporated into the host cell genome and is replicated along with the host genome. The virus may survive in this way for many host cell generations. Sooner or later, an environmental signal can cause the host cell to begin producing virions—at which point the viral reproductive cycle enters the lytic phase.
LINK
The different types of viruses are described in Concept 19.4
FIGURE 11.7 illustrates molecular events in the lytic life cycle of T4, a typical double-stranded DNA bacteriophage (phage, or bacterial virus). At the molecular level, the lytic cycle has two stages, early and late:
- The viral genome contains a promoter that binds host RNA polymerase. In the early stage, viral genes that lie adjacent to this promoter are transcribed. These early genes encode proteins that shut down expression of host genes, stimulate viral genome replication, and activate the transcription of viral late genes. The host genes are shut down by a posttranscriptional mechanism: a virus-encoded enzyme degrades the host RNA before it can be translated. Another viral nuclease digests the host’s chromosome, providing nucleotides for the synthesis of many copies of the viral genome. These processes can occur within a few minutes after the virus first infects the cell.
- In the late stage, viral late genes are transcribed; they encode the viral capsid proteins and enzymes that lyse the host cell to release the new virions.
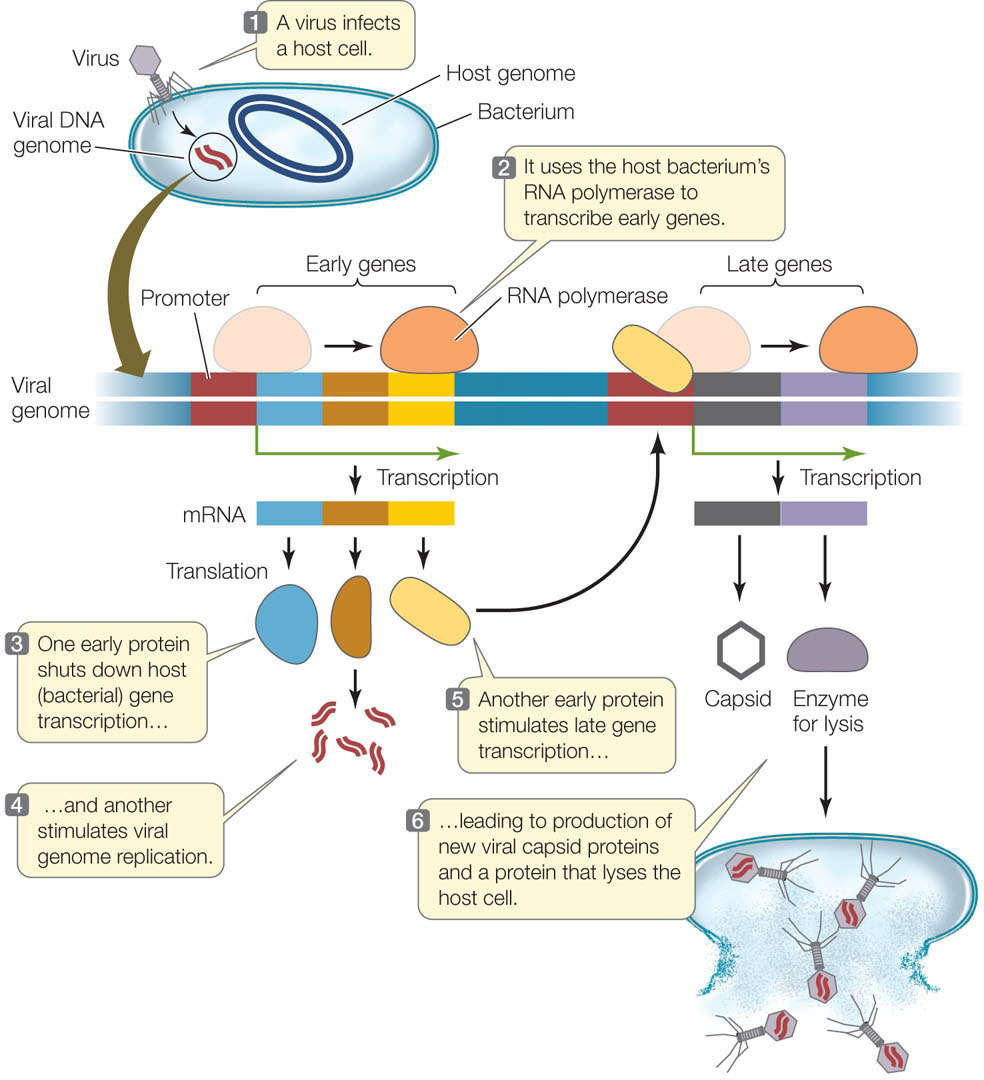
Under ideal conditions, this entire process—from binding and infection to release of new phage—can be completed in only half an hour.
CHECKpoint CONCEPT 11.1
- What is the difference between positive and negative regulation of gene expression?
- Describe the molecular conditions at the lac operon promoter in the presence and absence of lactose.
- Describe the molecular events at the trp operon promoter in the presence and absence of tryptophan.
- If the lac repressor gene were mutated so that the allosteric site on the encoded protein no longer bound allolactose, what would be the effect on transcription of the lac operon? What about a similar mutation in the trp repressor gene?
- What would be the effect of a mutation in the gene that encodes RNA polymerase so that it does not bind to the late gene promoter of bacteriophage T4?
Studies of bacteria and bacteriophage provide a basic understanding of the mechanisms that regulate gene expression and of the roles of regulatory proteins in both positive and negative regulation. We will now turn to the control of gene expression in eukaryotes. You will see both negative and positive control of transcription, as well as posttranscriptional mechanisms of regulation.
222