Concept 11.3: Gene Expression Can Be Regulated via Epigenetic Changes to Chromatin
So far we have focused on regulatory events that involve specific DNA sequences at or near a gene’s promoter. Eukaryotic cells are also able to regulate transcription via reversible, non-sequence-specific alterations to either the DNA or the chromosomal proteins that package the DNA in the nucleus. These alterations can be passed on to daughter cells after mitosis or meiosis. They are called epigenetic changes to distinguish them from mutations, which involve irreversible changes to the DNA’s base sequence (see Concept 9.3).
Modification of histone proteins affects chromatin structure and transcription
Epigenetic gene regulation can occur via the alteration of chromatin structure, or chromatin remodeling. Large amounts of DNA (nearly 2 meters in humans!) are packed within the nucleus (which has a diameter of about 5 μm). The basic unit of DNA packaging in eukaryotes is the nucleosome, a core of positively charged histone proteins around which DNA is wound:
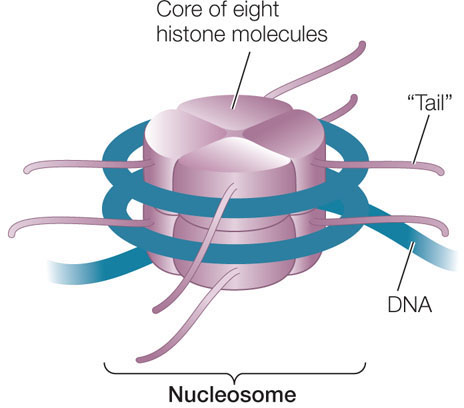
Each histone protein has a “tail” of approximately 20 amino acids at its N terminus that sticks out of the compact structure and contains certain positively charged amino acids, notably lysine. Ordinarily there is strong ionic attraction between the positively charged histone proteins and DNA, which is negatively charged because of its phosphate groups. Because of this attraction, nucleosomes can make DNA physically inaccessible to RNA polymerase and the rest of the transcription apparatus. However, a variety of transcription factors and RNA polymerase can bind enzymes called histone acetyltransferases, which add acetyl groups to these positively charged amino acids and neutralize their charges:
227
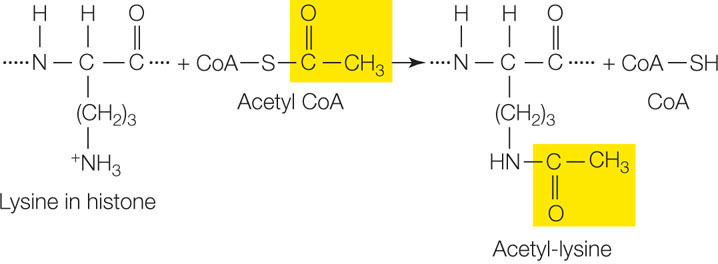
Reducing positive charges on the histone tails reduces the affinity of the histones for the DNA, loosening the compact nucleosome (FIGURE 11.14). The majority of histone acetylation is found near gene promoters, but acetylated histones are also found throughout the transcribed regions of genes. Thus histone acetylation promotes both transcription initiation and elongation. Histone acetylases can be recruited to promoters by transcription factors. An example is CREB, the transcription factor that is associated with addiction (see the opening story of this chapter). CREB binds specific acetyltransferases that participate in the activation of CREB-responsive genes.
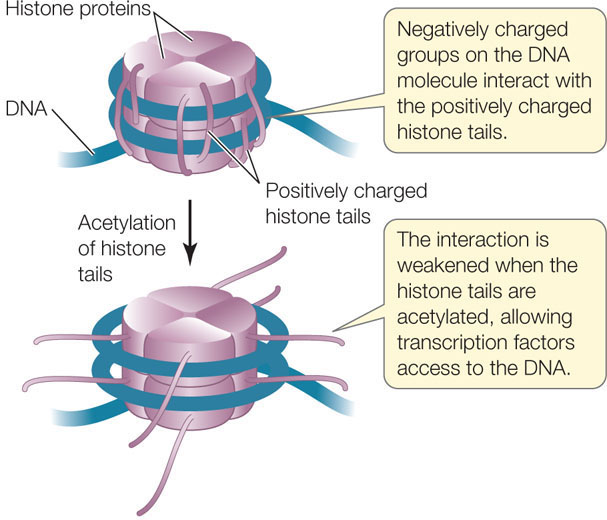
Another class of chromatin remodeling proteins, histone deacetylases, can remove the acetyl groups from histones and thereby repress transcription. Histones can also be modified in other ways, including methylation (the addition of a methyl group) and phosphorylation (the addition of a phosphate group). Histone methylation can contribute to either the activation or repression of gene expression, depending on which lysine residue is methylated. Histone phosphorylation is involved in chromosome condensation during mitosis and meiosis, as well as affecting gene regulation. All of these effects are reversible, and so the transcriptional activity of a eukaryotic gene may be determined by varying patterns of histone modification.
DNA methylation affects transcription
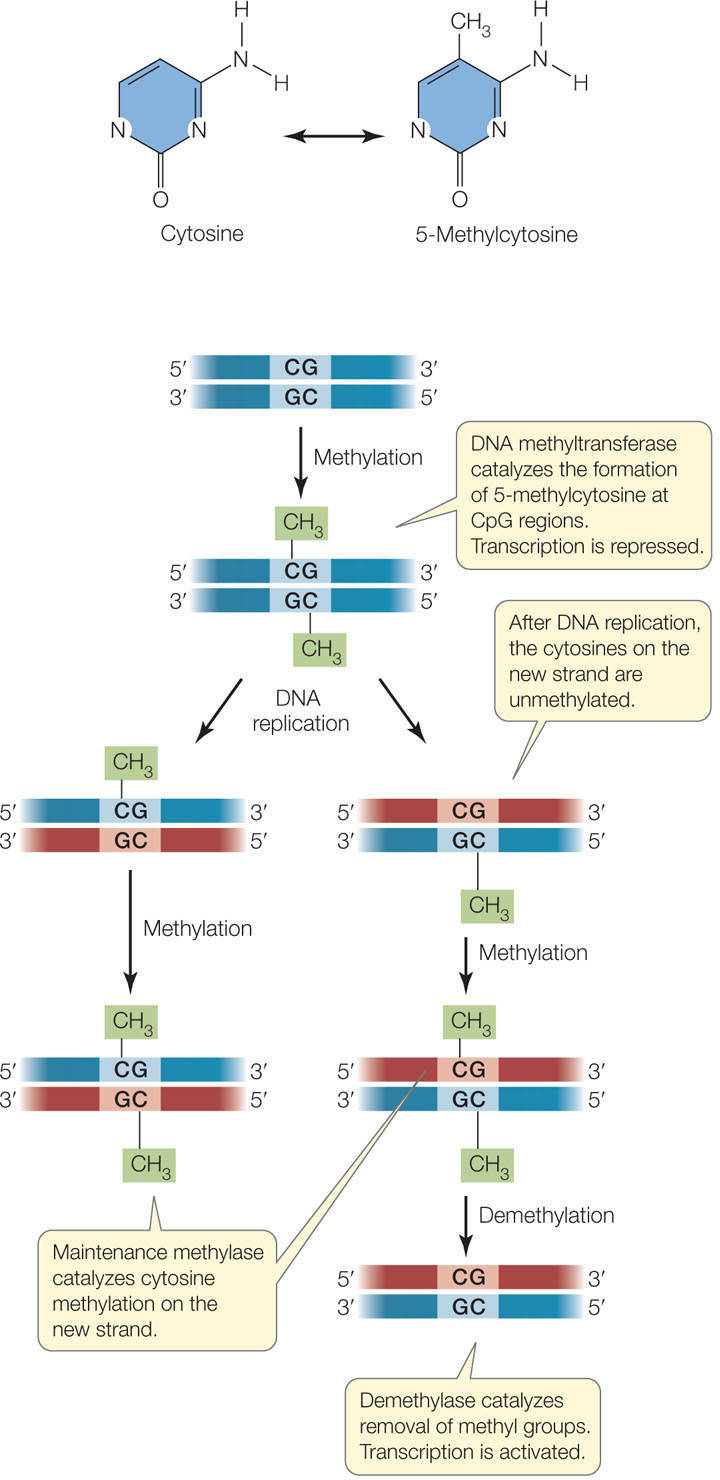
Depending on the organism, from 1 to 5 percent of cytosines in the DNA are chemically modified by the addition of a methyl group (—CH3), to form 5-methylcytosine (FIGURE 11.15). This covalent addition is catalyzed by the enzyme DNA methyltransferase and, in mammals, usually occurs on cytosines (C) that are adjacent to guanines (G). Virtually all of these CpG (“p” is for the phosphate in the DNA backbone) sites are methylated, apart from those found in and near transcriptionally active promoters. Promoters usually contain regions of DNA that are rich in CpG sites, and such regions are called CpG islands.
228
Methylated DNA binds specific proteins that are involved in the repression of transcription; thus heavily methylated genes tend to be inactive (silenced). Whereas histone acetylation/deacetylation (see above) are dynamic processes resulting in short-term changes in gene expression, DNA methylation is usually a stable, long-term silencing mechanism. When DNA is replicated, a maintenance methyltransferase catalyzes the formation of 5-methylcytosine in the new DNA strands. However, the pattern of cytosine methylation can also be altered, because methylation is reversible: a third enzyme, appropriately called demethylase, catalyzes the removal of the methyl group from cytosine (see Figure 11.15). In ways that are not fully understood, the enzymes involved in histone modification and DNA methylation/demethylation interact to ensure that genes whose products are needed in the cell are kept unmethylated, and their associated histones acetylated.
Sometimes, large stretches of DNA or almost entire chromosomes are methylated. Under a microscope, two kinds of chromatin can be distinguished in the stained interphase nucleus: euchromatin and heterochromatin. The euchromatin appears diffuse and stains lightly; it contains the DNA that is transcribed into mRNA. Heterochromatin is condensed and stains darkly; any genes it contains are generally not transcribed.
A dramatic example of heterochromatin is the X chromosome in female mammals. A normal female mammal has two X chromosomes, whereas a normal male has an X and a Y (see Concept 8.3). Because each female cell has two copies of each X chromosome gene, the female should have the potential to produce twice as much of each protein product as the male. Nevertheless, for 75 percent of the genes on the X chromosome, the total amount of mRNA produced is generally the same in males and in females. How does this happen?
In the early female embryo, one copy of X becomes hetero-chromatic and transcriptionally inactive in each cell, and the same X remains inactive in all of that cell’s descendants. In a given female embryo cell, the “choice” of which X to inactivate is random. Recall that one X in a female comes from her father and one from her mother. Thus in one embryonic cell the paternal X might be inactivated, but in a neighboring cell the maternal X might be inactivated.
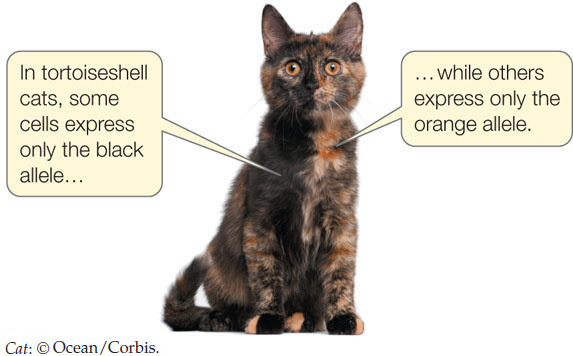
A familiar example of a phenotype caused by X chromosome inactivation is the tortoiseshell cat. In cats, two alleles of an X-linked gene that contributes to coat color are orange (XB) and black (Xb). In the embryo of a heterozygous female (XBXb), there is random X-inactivation such that in some cells only the orange allele is expressed, while in others it is the black allele. These cells then form the progenitors of patches of skin in the animal.
The inactive X is identifiable within the nucleus as a hetero-chromatic Barr body (named for its discoverer, Murray Barr):
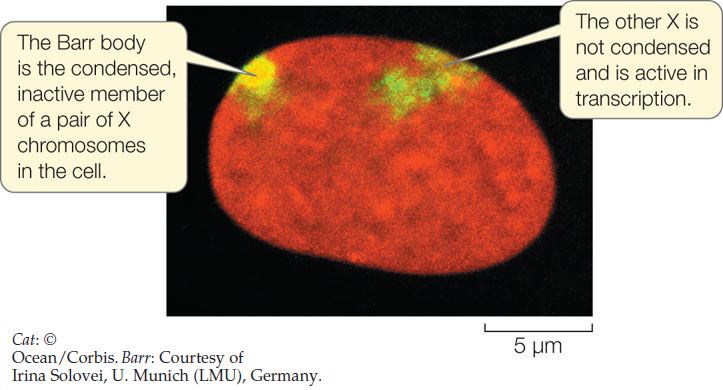
In this micrograph, both X chromosomes are stained yellow-green; the Barr body, which consists of heavily methylated DNA, is more condensed than the active X and therefore appears brighter.
Having the right “dosage” of transcriptionally active genes is important. This is illustrated by the fact that aneuploidy—an unusual number of a particular chromosome—is a harmful condition that often results in embryo death (see Concept 7.4).
Epigenetic changes can be induced by the environment
Methylation patterns are stable and can be passed on from one generation to the next. However, a recent study of human monozygotic (identical) twins shows that these patterns can be altered over time.
Monozygotic twins come from a single fertilized egg that divides to produce two separate cells; each of these develops into a separate individual. Identical twins thus have generally identical genomes. But are they identical in their “epigenomes”? A comparison of DNA in hundreds of such twin pairs shows that in tissues of 3-year-olds, DNA methylation patterns are virtually the same. But by age 50—when twins have usually been living apart and in different environments for decades—the patterns are quite different, and different genes are expressed. This indicates that the environment plays an important role in epigenetic modifications, and therefore in the regulation of genes that these modifications affect.

Go to MEDIA CLIP 11.1 The Surprising Epigenetics of Identical Twins
PoL2e.com/mc11.1
229
What factors in the environment lead to epigenetic changes? Chemicals such as tobacco smoke and dietary components such as folic acid can affect DNA methylation patterns. Another factor might be stress: when mice are put in a stressful situation, genes that are involved in important brain pathways become heavily methylated (and transcriptionally inactive). Treatment of the stressed mice with an antidepressant drug reverses these changes.
LINK
Stressful experiences can have lifelong effects on behavior patterns; see Concept 40.2
DNA methylation can result in genomic imprinting
In mammals, specific patterns of methylation develop for each sex during gamete formation. This happens in two stages: first, the existing methyl groups are removed from the 5′-methylcytosines by a demethylase, and then a DNA methylase adds methyl groups to a new set of cytosines. When the gametes form, they carry this new pattern of methylation.
The DNA methylation pattern in male gametes (sperm) differs from that in female gametes (eggs) at about 200 genes in the mammalian genome. That is, a given gene in this group may be methylated in eggs but unmethylated in sperm. In this case the offspring would inherit a maternal gene that is transcriptionally inactive (methylated) and a paternal gene that is transcriptionally active (demethylated). This is called genomic imprinting.
Imprinting of specific genes occurs primarily in mammals and flowering plants. Most imprinted genes are involved with embryonic development. An embryo must have both the paternally and maternally imprinted gene patterns to develop properly. In fact, attempts to make an embryo that has chromosomes from only one sex (for example, by chemically treating an egg cell to double its chromosomes) usually fail. So imprinting has an important lesson for genetics: males and females may be the same genetically (except for the X and Y chromosomes), but they differ epigenetically.
CHECKpoint CONCEPT 11.3
- What is the difference between epigenetic regulation and gene regulation by transcription factors?
- How can a DNA methylation pattern be inherited?
- In colorectal cancer, some tumor suppressor genes are inactive. This is an important factor resulting in uncontrolled cell division. Two of the possible explanations for the inactive genes are: (1) a mutation in the coding region, resulting in an inactive protein, and (2) epigenetic silencing at the promoter of the gene, resulting in reduced transcription. How would you investigate each of these possibilities?
Thus far we have examined transcriptional gene regulation in viruses, prokaryotes, and eukaryotes. In the final concept we will focus on the posttranscriptional mechanisms for regulating gene expression in eukaryotes.