Concept 13.2: DNA Can Genetically Transform Cells and Organisms
One goal of recombinant DNA technology is to clone—produce many identical copies of—a particular DNA sequence. We have seen the term “clone” used in the context of whole cells or organisms that are genetically identical to one another (see Concept 7.1). A gene can be cloned by inserting it into a bacterial cell such as E. coli. The bacterium is allowed to reproduce and multiply into millions of identical cells, all carrying copies of the gene. Cloning might be done for sequence analysis, to produce a protein product in quantity, or as a step toward creating an organism with a new phenotype.
The process of inserting recombinant DNA into host cells is called transformation, or transfection if the host cells are derived from an animal. A host cell or organism that contains recombinant DNA is described as transgenic. Later in this chapter we will encounter many examples of transgenic cells and organisms, including yeast, mice, rice plants, and even cattle.
Various methods are used to create transgenic cells. Generally, only a few of the cells that are exposed to the recombinant DNA actually become transformed with it. In order to grow only the transgenic cells, selectable marker genes, such as genes that confer resistance to antibiotics, are often included as part of the recombinant DNA molecule. Antibiotic resistance genes were the markers used in Cohen and Boyer’s experiment (see Figure 13.4).
Genes can be inserted into prokaryotic or eukaryotic cells
In theory, any cell or organism can act as a host for the introduction of recombinant DNA. Most research has been done using model organisms:
- Bacteria are easily grown and manipulated in the laboratory. Much of their molecular biology is known, especially for well-studied bacteria such as E. coli. Furthermore, bacteria contain plasmids, which are easily manipulated to carry recombinant DNA into the cell. Because the processes of transcription and translation proceed differently in prokaryotes than they do in eukaryotes, however, bacteria might not be suitable as hosts to express eukaryotic genes.
- Yeasts such as Saccharomyces are commonly used as eukaryotic hosts for recombinant DNA studies. The advantages of using yeasts include rapid cell division (a life cycle completed in 2 hours), ease of growth in the laboratory, and a relatively small genome size. In addition, yeasts have most of the characteristics of other eukaryotes, except for those characteristics involved in multicellularity.
- Plant cells are good hosts, because even fully differentiated plant cells can be treated with hormones that make them dedifferentiate into unspecialized stem cells (see Concept 13.3). The unspecialized cells can be transformed with recombinant DNA and then studied in culture, or grown into new plants. There are also methods for making whole transgenic plants without going through the cell culture step. These methods result in plants that carry the recombinant DNA in all their cells, including the germline cells.
- Cultured animal cells can be used to study the expression of human or animal genes, for example for medical purposes. Whole transgenic animals can also be created.
A variety of methods are used to insert recombinant DNA into host cells
Methods for inserting DNA into host cells vary. The cells may be chemically treated to make their outer membranes more permeable, and then mixed with the DNA so it can diffuse into the cells. Another approach is called electroporation: a short electric shock is used to create temporary pores in the membranes through which the DNA can enter. Viruses can be altered so that they carry or insert recombinant DNA into cells. A common method for transforming plants involves a specific bacterium that inserts DNA into cells of some plants. Transgenic animals can be produced by injecting recombinant DNA into the nuclei of fertilized eggs. There are even “gene guns,” which “shoot” the host cells with tiny particles carrying the DNA.
259
The challenge of inserting new DNA into a cell lies not just in getting it into the host cell, but in getting it to replicate as the host cell divides. DNA polymerase does not bind to just any sequence. If the new DNA is to be replicated, it must become part of a segment of DNA that contains an origin of replication (see Concept 9.2). Such a DNA molecule is called a replicon, or replication unit.
There are two general ways in which the newly introduced DNA can become part of a replicon within the host cell:
- It may be inserted into a host chromosome. Although the site of insertion is usually random, this is nevertheless a common method of integrating new genes into host cells.
- It can enter the host cell as part of a carrier DNA sequence, called a vector, and can either integrate into the host chromosome or have its own origin of DNA replication.
Several types of vectors are used to get DNA into cells.
Plasmids As Vectors
As we described in Concept 8.4, plasmids are small, circular DNA molecules that replicate autonomously in many prokaryotic cells. A number of characteristics make plasmids useful as transformation vectors:
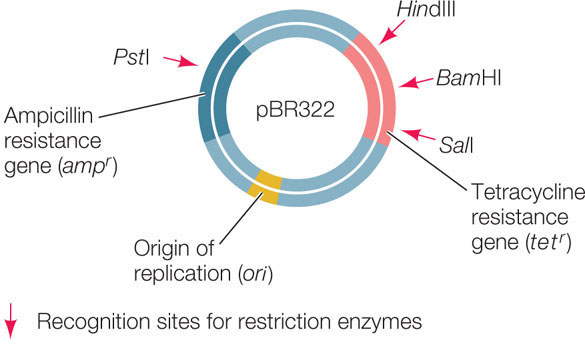
- Plasmids are relatively small (an E. coli plasmid usually has 2,000–6,000 bp) and are therefore easy to manipulate in the laboratory.
- A typical plasmid has one or more restriction enzyme recognition sequences that each occur only once in the plasmid sequence. These sites make it easy to insert additional DNA into the plasmid before it is used to transform host cells.
- Many plasmids contain genes that confer resistance to antibiotics and thus can serve as selectable markers.
- Plasmids have a bacterial origin of replication (ori) and can replicate independently of the host chromosome. It is not uncommon for a bacterial cell to contain hundreds of copies of a recombinant plasmid. For this reason, the power of bacterial transformation to amplify a gene is extraordinary. A 1-liter culture of bacteria harboring the human β-globin gene in a typical plasmid can have as many copies of that gene as there are cells in a typical adult human (1014).
The plasmids used as vectors in the laboratory have been extensively altered to include convenient features: multiple cloning sites with 20 or more unique restriction enzyme sites for cloning purposes; origins of replication for a variety of host cells; and various kinds of reporter genes and selectable marker genes. An example is pBR322, a plasmid used to transform E. coli:
Plasmid Vectors for Plants
An important vector for carrying new DNA into many types of plants is a plasmid found in the bacterium Agrobacterium tumefaciens. This bacterium lives in the soil, infects plants, and causes a disease called crown gall, which is characterized by the presence of growths (or tumors) on the plant. A. tumefaciens contains a plasmid called Ti (for tumor-inducing). When the bacterium infects a plant cell, a region of the Ti plasmid called the T DNA is inserted into the cell, where it becomes incorporated into one of the plant’s chromosomes. The Ti plasmid carries the genes needed for this transfer and incorporation of the T DNA:
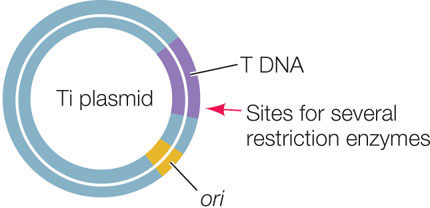
The T DNA carries genes that are expressed by the host cell, causing the growth of tumors and the production of specific sugars that the bacterium uses as sources of energy. Scientists have exploited this remarkable natural “genetic engineer” to insert foreign DNA into the genomes of plants. When used as a vector for plant transformation, the tumor-inducing and sugar-producing genes on the T DNA are removed and replaced with foreign DNA. The altered Ti plasmids are first used to transform Agrobacterium cells from which the original Ti plasmids have been removed. Then the Agrobacterium cells are used to infect plant cells.
Viruses As Vectors
Constraints on plasmid replication limit the size of the new DNA that can be inserted into a plasmid to about 10,000 bp. Although many prokaryotic genes may be smaller than this, most eukaryotic genes—with their introns and extensive flanking sequences—are bigger. A vector that accommodates larger DNA inserts is needed for these genes.
Both prokaryotic and eukaryotic viruses are often used as vectors for eukaryotic DNA. Bacteriophage λ, which infects E. coli, has a DNA genome of about 45,000 bp; this is all that fits into the phage head. If the phage genes that cause the host cell to die and lyse—about 20,000 bp—are eliminated, the virus can still attach to a host cell and inject its DNA, but the host cell will not die. The deleted 20,000 bp can be replaced with DNA from another organism. Because viruses infect cells naturally, they offer a great advantage over plasmids, which often require artificial means to coax them to enter host cells.
260
APPLY THE CONCEPT: DNA can genetically transform cells and organisms
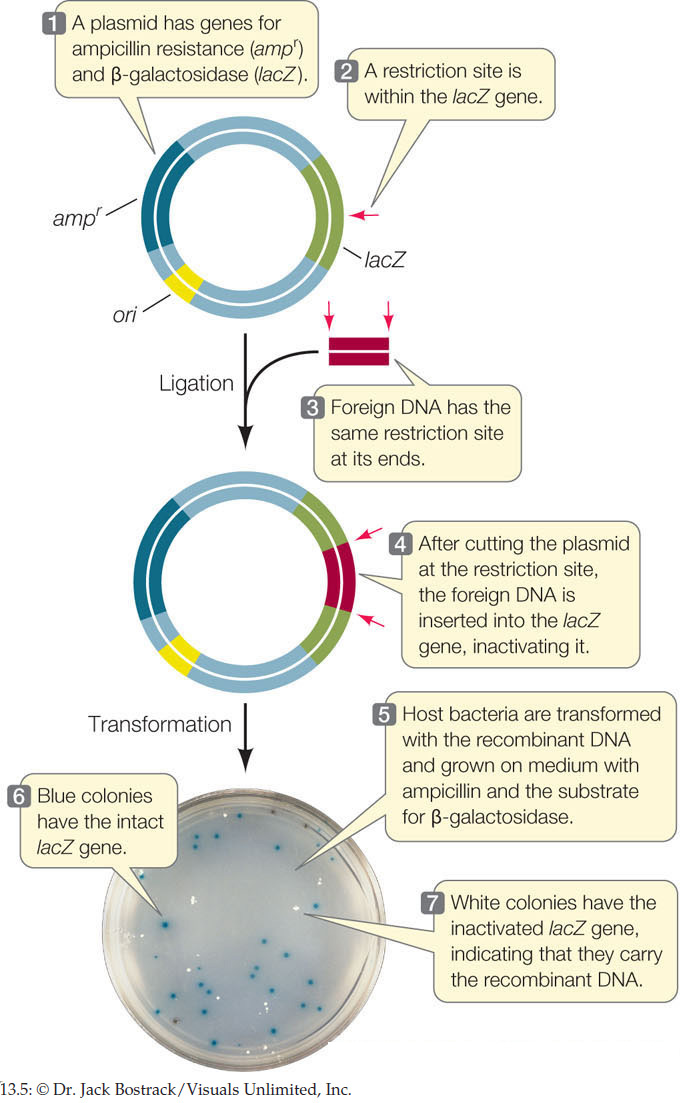
As shown in Figure 13.5, the β-galactosidase (lacZ) gene encodes an enzyme that can convert the colorless substrate X-gal into a bright blue product. A plasmid vector contains a modified version of the lacZ gene with multiple restriction sites within its coding sequence, and a gene for resistance (R) to the antibiotic ampicillin. This vector is used with an E. coli strain that carries no other functional lacZ gene and that is sensitive (S) to ampicillin. A biologist clones a wheat gene by inserting it into the multiple cloning site of the lacZ gene in this vector. As a control to ensure that the bacterial cells are viable, she also grows some cells that were not transformed with the ligation products. Remember that after a ligation reaction, only a few of the plasmids are recombinant. Fill in the table with the results of these transformations.

Reporter genes help select or identify host cells containing recombinant DNA
Even when a population of host cells interacts with an appropriate vector, only a small proportion of the cells actually take up the vector. Furthermore, the process of making recombinant DNA is far from perfect. After a ligation reaction, not all the vector copies contain the foreign DNA. How can we identify or select the host cells that contain the vector with foreign DNA?
As we described above, selectable markers such as antibiotic resistance genes can be used to select cells containing those genes. These cells can be selected because only cells carrying the antibiotic resistance gene can grow in the presence of that antibiotic. Selectable markers are one type of reporter gene, which is any gene whose expression is easily assayed. Other reporter genes code for proteins that can be detected visually. Two commonly used reporter genes are lacZ and the gene for green fluorescent protein:
- The β-galactosidase (lacZ) gene (see Figure 11.3) codes for an enzyme that can convert the white substrate X-gal into a bright blue product. Many cloning plasmids contain the lacZ gene, along with genes for antibiotic resistance. As shown in FIGURE 13.5, foreign DNA can be inserted into the lacZ gene, inactivating it. Bacteria transformed with the plasmid are selected on a solid medium containing X-gal and the appropriate antibiotic. Clones containing the recombinant plasmid cannot make β-galactosidase, and produce white colonies. Clones that contain the original plasmid with no insert express the lacZ gene and make blue colonies.
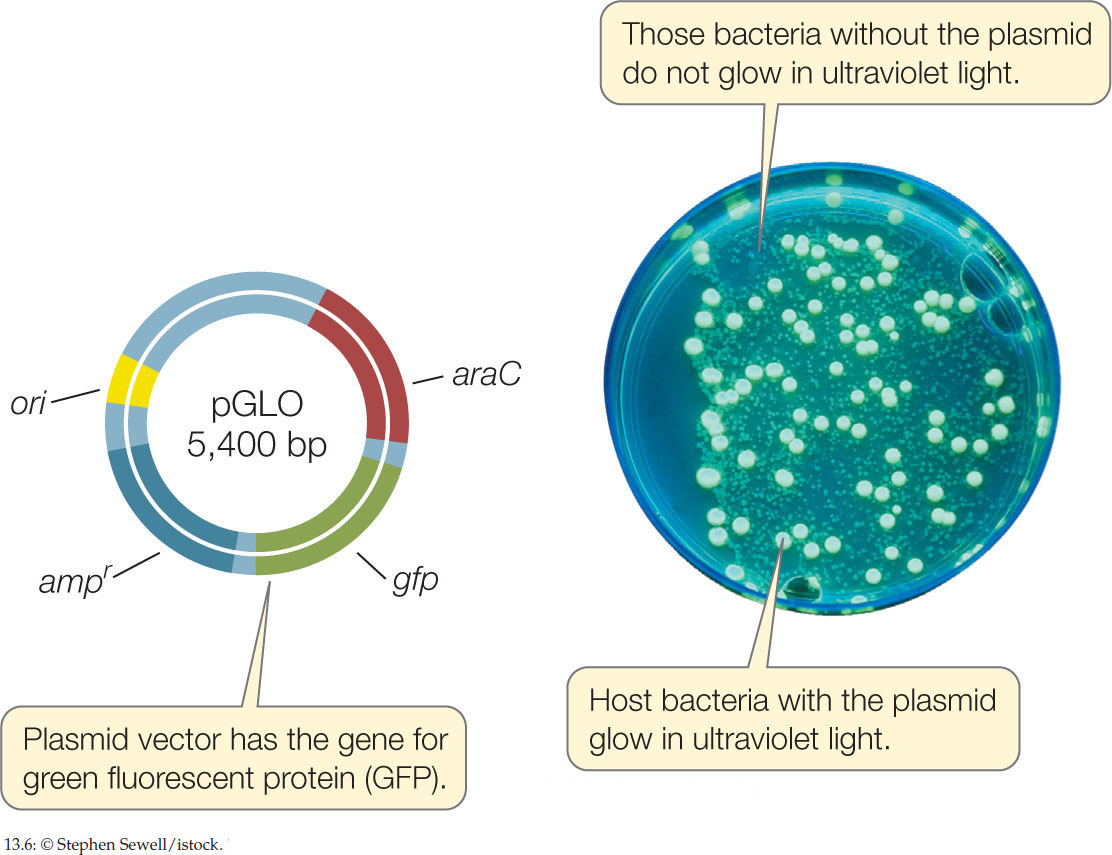
- Green fluorescent protein (GFP), which normally occurs in the jellyfish Aequorea victoria, emits green light when exposed to ultraviolet light. The gene for this protein is now widely used as a reporter gene (FIGURE 13.6).
RESEARCH TOOLS
LINK
The lacZ gene is part of the inducible lac operon in E. coli, which encodes proteins required for β-galactoside metabolism; see Concept 11.1
261
Such reporters are not just used to select and identify cells carrying recombinant DNA. They can be attached to promoters in order to study how the promoters function under different conditions or in different tissues of a transgenic multicellular organism. Reporters can also be attached to other proteins, to study how and where those proteins become localized within eukaryotic cells.
CHECKpoint CONCEPT 13.2
- Outline the steps used to create recombinant DNA, transform host cells, and detect cells carrying the recombinant DNA.
- “Shuttle vectors” have the ability to transform both prokaryotic and eukaryotic cells. What sequences would you expect these vectors to have?
- What are the advantages of green fluorescent protein over antibiotic resistance as a marker on a plasmid for genetic transformation?
- Plasmid X has ampicillin and tetracycline resistance genes. The restriction enzyme EcoRI cleaves the plasmid once, within the tetracycline resistance gene. Plasmid B has a streptomycin resistance gene and one site for EcoRI cleavage that is not within the resistance gene. The two plasmids are cut with EcoRI and treated with DNA ligase. The mixture is used to transform an E. coli strain that is sensitive to the three antibiotics. Which antibiotic(s) would you add to the bacterial growth medium to select those bacteria carrying a recombinant plasmid?
We have described how DNA can be cut, inserted into a vector, and introduced into host cells. We have also seen how host cells carrying recombinant DNA can be identified. Now let’s consider the sources of DNA used for cloning, as well as some molecular methods for manipulating gene expression.