Concept 33.1: Muscle Cells Develop Forces by Means of Cycles of Protein–Protein Interaction
Muscle tissue makes up a large portion of body mass in vertebrates and many other animals. In humans, for example, muscles account for almost half of body mass (FIGURE 33.1). Muscles are as important functionally as they are anatomically. They are the basis for virtually all behavior. They permit animals to run, fly, or swim to acquire food or escape dangers. In humans and some other mammals, muscles of the face account for changing facial expressions. Besides behavior, many other physiological actions also depend on muscle contraction, such as the circulation of blood by the beating of the heart.

There are several types of muscle tissue in the animal kingdom. The contractile mechanism is basically the same in most muscle types, however. We will begin our study of muscle function by focusing on the contractile mechanism in vertebrate skeletal muscle—the type of muscle that is attached to the bones of the skeleton and that provides the power for walking, flying, swimming, and other forms of locomotion.
Contraction occurs by a sliding-filament mechanism
Muscle tissue develops mechanical forces. This is its defining feature. In the scientific study of muscle, contraction refers to force development. A muscle does not necessarily shorten when it undergoes contraction. To see the distinction between contraction and shortening, imagine grabbing a bar of steel attached to a concrete wall with your hand and pulling as hard as you can with your arm muscles. Your muscles will develop great forces, but they will not shorten because the bar of steel is immovable. For a muscle to shorten during contraction, the force it exerts must be greater than opposing forces. When you use your arm to pull on a drawer, your arm muscles shorten during contraction because the force they exert exceeds the opposing forces, and the drawer slides toward you.
The current theory of how muscle contracts is often known as the sliding-filament theory. This name describes what the process looks like and is useful in that way. When muscle cells contract in circumstances in which they shorten, tiny filaments in the cells look like they slide past each other during contraction. However, the name “sliding” can be misleading, because “slide” suggests effortless motion, like a sled sliding down a snow-covered hill. In fact, the motion of the filaments in muscle cells is not at all effortless. It is quite the opposite: during contraction, the filaments use energy from ATP to develop mechanical forces and pull on each other. The “sliding” of the filaments is merely a consequence of this energy-using, force-generating process.
Actin and myosin filaments slide in relation to each other during muscle contraction
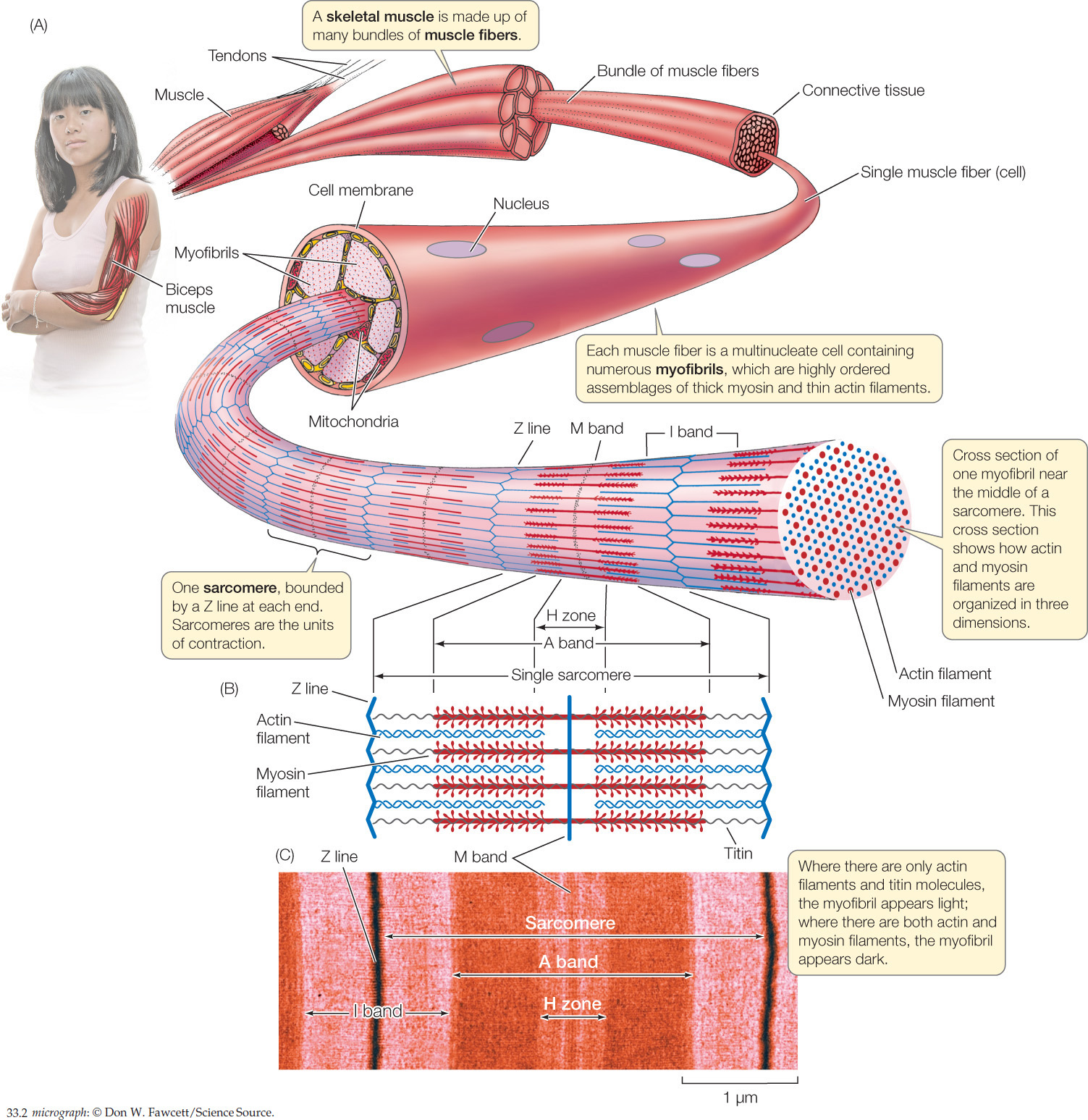
The cells of skeletal muscle are sometimes called “cells” and sometimes “fibers.” These two terms are synonyms: a muscle fiber is a muscle cell. The cells in vertebrate skeletal muscle are large and have many nuclei, making them multinucleate. During development they form through the fusion of many individual embryonic muscle cells called myoblasts. A muscle such as your biceps (the muscle that pulls your forearm up toward your upper arm) is composed of hundreds or thousands of muscle fibers that are organized in bundles surrounded by connective tissue (Figure 33.2A).
Go to ACTIVITY 33.1 The Structure of a Sarcomere
PoL2e.com/ac33.1
Muscle contraction results from the interaction of two contractile proteins that are abundant in each muscle fiber: actin and myosin. Molecules of actin are organized into actin filaments, also called thin filaments. Molecules of myosin are organized into myosin filaments, or thick filaments. These two kinds of filaments interdigitate in parallel arrays as seen in Figure 33.2B.
LINK
Actin and myosin are also part of the cytoskeleton, which is responsible for other types of movement in cells; see Concept 4.4
Let’s look now in more detail at the anatomical relationships among a muscle, its muscle fibers, and the actin and myosin filaments inside each fiber. Looking at Figure 33.2A and moving outward from the person’s arm, you will see that a muscle is made up of bundles of long, parallel muscle fibers. Similarly, each muscle fiber is composed internally of long, parallel myofibrils, each of which is composed of arrays of actin and myosin filaments oriented parallel to the long axis of the myofibril.
683
Understanding the structure of a myofibril is critical. Figure 33.2A is drawn so you can view a myofibril from the side. From this view, you can see that a myofibril consists of repeating units called sarcomeres. Each sarcomere starts and ends with a Z line. Internally, as shown in Figure 33.2B, each sarcomere contains overlapping arrays of actin and myosin filaments, as we have already noted. At the far right of Figure 33.2A, the myofibril is shown in cross section so you can see the three-dimensional arrangement of its actin and myosin filaments. In most regions of a myofibril, as the cross section shows, each thick myosin filament is surrounded by six thin actin filaments, and each actin filament sits within a triangle of three myosin filaments.
In side view, the structures we have been discussing give skeletal muscle a distinctly banded appearance when the muscle is examined under a simple light microscope. To see individual actin and myosin filaments, you would need to use an electron microscope because they are far too thin to be seen with a light microscope. When you look at muscle with a light microscope, however, you see patterns that result from the organization of large sets of actin and myosin filaments—much as you can see black and white lines on a computer screen even though you cannot see the individual pixels that collectively give rise to the patterns. Figure 33.2C shows the banding pattern in a single muscle fiber as seen with a light microscope at fairly high magnification. By comparing Figures 33.2B and 33.2C, you can see how the bands seen with the light microscope line up with the actin and myosin filaments in a sarcomere. At lower magnification under a light microscope, the banding in a region of skeletal muscle looks like this:
684

Because of this banding pattern, skeletal muscle is called striated muscle.
Before the molecular nature of the muscle banding pattern was known, the bands were given names that are still used today (see Figure 33.2B,Figure 33.2C). Each sarcomere is bounded by Z lines, as already mentioned, which represent structures to which the actin filaments are attached. At the center of each sarcomere is a structure called the M band (for “middle”). It contains proteins that help hold the myosin filaments in position. Each myosin filament extends out symmetrically on either side of the M band. Collectively, the myosin filaments in each sarcomere give rise to a dark region—called the A band—centered on the M band.
Finally, in each sarcomere there are three relatively light bands: an H zone centered on the M band, and one half of an I band at each end of the sarcomere. These bands appear light because, unless a myofibril is fully contracted, the actin and myosin filaments do not overlap in them: only myosin filaments are found in the H zone, and only actin filaments are found in the I bands (see Figure 33.2B,Figure 33.2C).
Within a sarcomere, the thick myosin filaments are held in their correct positions in three-dimensional space by molecules of a protein called titin (Figure 33.3A). This giant molecule is the largest known protein. A myosin filament consists of about 200–400 individual myosin molecules in a bundle. Each titin molecule in a sarcomere runs between the M band and a Z line, directly through a myosin filament along the filament’s long axis. Between the end of the filament closest to the Z line and the Z line itself, the titin molecule is elastic, like a bungee cord. Collectively, the titin molecules do two things. They hold the myosin filaments in position, and if a sarcomere becomes stretched, their elasticity returns the sarcomere to its shorter length when the stretching forces relax.

A myofibril looks like Figure 33.3A when it is in its relaxed state. When a myofibril contracts in circumstances in which it can shorten, its sarcomeres shorten and the banding pattern changes to look like Figure 33.3B. The light-colored H zone and I bands become much narrower, and the dark Z lines move toward the A band. These changes are easy to see under a light microscope as Figure 33.3 shows. Why do they occur? At a molecular level, during contraction the actin and myosin filaments slide past each other, so that they overlap to an increased degree, shortening the muscle fiber. In each sarcomere the actin filaments slide into the H zone, the region occupied by only myosin filaments in the relaxed state. As they do so, the Z lines at opposite ends of the sarcomere are pulled closer to each other.
ATP-requiring actin–myosin interactions are responsible for contraction
To understand how a sarcomere develops mechanical forces during contraction, it is critical to understand the structures of myosin and actin molecules (FIGURE 33.4). A myosin molecule consists of two long polypeptide chains coiled together, each ending in a large globular head. A myosin filament is made up of many myosin molecules, the heads of which project sideways near the ends of the filament.


Go to ANIMATED TUTORIAL 33.1 Molecular Mechanisms of Muscle Contraction
PoL2e.com/at33.1
685
An actin filament consists of small actin monomers polymerized into two long chains that look like two strands of pearls twisted together. Twisting around each actin chain is another protein, tropomyosin, and attached to tropomyosin at intervals are molecules of still another protein, troponin. These molecules control contraction and relaxation, as we will see in the next section.
How do myosin and actin interact? The globular heads of myosin can bind to specific sites on actin, forming bridge-like links, called cross-bridges, between the myosin and actin filaments. Two functional properties of the myosin heads are exceedingly important. First, when a myosin head binds to an actin filament, the head’s molecular shape (conformation) changes in such a way that the head bends and exerts a tiny force on the actin filament. This force causes the actin filament to be pulled a distance of 5–10 nanometers toward the M band along the myosin filament (see the transition from step 3 to step 4 in Figure 33.7). Second, when a myosin head is bound to actin, it has ATPase activity: that is, it binds to and hydrolyzes ATP, releasing energy. The energy released changes the molecular shape of the myosin head, causing it to disconnect from actin and unbend, returning to its extended position. At that point, the myosin head can again bind to actin and undergo another change of molecular shape that causes a tiny force to be exerted on the actin filament, moving the actin filament relative to the myosin filament.
Together, these details explain how the actin and myosin filaments in a sarcomere interact to develop mechanical forces and slide past each other during contraction. The details also help explain why the force a muscle fiber can exert during contraction depends on how many actin–myosin cross-bridges can form.
Rigor mortis—the stiffening of muscles soon after death—is still another muscle property that can be explained by the details of muscle contraction we are discussing. ATP is needed to break actin–myosin bonds. When ATP production ceases with death, actin–myosin bonds cannot be broken and muscles stiffen.
To fully understand contraction, remember that each myosin filament has many myosin heads near both ends and is surrounded by six actin filaments. As contraction takes place, numerous myosin heads go through cycles of interaction with the actin filaments. Their cycles of binding and release are not exactly synchronous, however. Accordingly, when a single myosin head breaks its contact with actin, other cross-bridges are still connected and flexing. As a result, the actin filaments do not slip backward.
Excitation leads to contraction, mediated by calcium ions
Muscle cells, as well as nerve cells (see Chapter 34), are excitable. A cell is excitable if its cell membrane can generate and conduct electrical impulses (action potentials). Cell membranes ordinarily have an electrical polarity in which the outside of the membrane is more positive than the inside. An impulse, or action potential, is a region of reversed polarity, also called depolarization (see Concept 34.2). In an excitable cell, after an impulse—a region of reversed polarity, or depolarization—is initiated at one point in a cell membrane, it travels along the full length of the cell membrane:
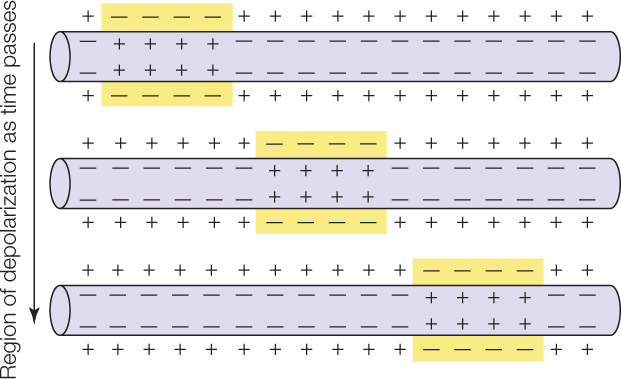
What triggers muscle cells to contract? In skeletal muscle, nerve impulses stimulate muscle fibers to contract. The nerve cells that are involved have long, threadlike cell processes (cell extensions) called axons (see Figure 34.2). Each muscle fiber is in physical contact with an axon of a nerve cell. The nerve cell is said to innervate the muscle fiber. The point of contact between the nerve cell and muscle fiber is known as a neuromuscular junction (FIGURE 33.5). When a nerve impulse arrives at the neuromuscular junction, it is said to excite the muscle fiber, and the event is called excitation. During excitation (see Concept 34.3), an impulse or action potential is initiated in the cell membrane of the muscle fiber and then travels the length of that cell membrane.

686
Excitation–contraction coupling is the process by which the excitation of a muscle fiber leads to contraction of the muscle fiber. Several decades ago, when excitation–contraction coupling was first being investigated, researchers injected muscle fibers in a frog’s leg with the bioluminescent protein aequorin, which is activated to emit light by calcium ions (Ca2+). The researchers found that these injected fibers emitted a flash of light each time they were stimulated to contract. This elegant experiment announced to the world that Ca2+ plays a role in excitation–contraction coupling. After a muscle fiber is excited, Ca2+ is released into the general cytoplasm of the fiber from internal Ca2+-storage locations. Ca2+ in the cytoplasm of an injected cell activates the aequorin, producing a flash of light.
Where is the Ca2+ stored in muscle fibers, and how is it released? A distinctive feature of muscle cells is that each cell has many spots where the cell membrane is indented inward into the cell cytoplasm to form a transverse tubule, or T tubule (FIGURE 33.6). The membrane that forms the walls of each T tubule is continuous with the outer cell membrane. Thus when an impulse or action potential travels along the outer cell membrane, it also travels inward along each T tubule, thereby spreading into the interior of the cell at multiple places via the system of multiple T tubules.

Go to ACTIVITY 33.2 The Neuromuscular Junction
PoL2e.com/ac33.2
The T tubules of a muscle cell are physically very close to the endoplasmic reticulum of the cell. In a muscle cell, the cytoplasm is called the sarcoplasm, and the endoplasmic reticulum is called the sarcoplasmic reticulum. The sarcoplasmic reticulum is a closed compartment that surrounds every myofibril in a muscle cell (see Figure 33.6). Calcium pumps in the sarcoplasmic reticulum membrane take up Ca2+ from the sarcoplasm. Thus when a muscle cell is at rest there is a high concentration of Ca2+ inside the sarcoplasmic reticulum and a low concentration in the sarcoplasm. Essentially, Ca2+ is stored inside the sarcoplasmic reticulum when a muscle cell is resting.
687
Spanning the space between the membranes of the T tubules and the membranes of the sarcoplasmic reticulum are two proteins that are physically connected. One of these, the dihydropyridine (DHP) receptor, is located in the T tubule membrane. It changes its conformation—its molecular shape—when an action potential reaches it. The other protein, the ryanodine receptor, is a Ca2+ channel located in the sarcoplasmic reticulum membrane.
The two receptor proteins act to release Ca2+ into the sarcoplasm when a muscle fiber is excited. When the DHP receptor is activated by an action potential traveling down a T tubule, both the DHP and ryanodine receptor proteins change conformation. As a consequence of this change, the ryanodine receptor (a Ca2+ channel) allows Ca2+ to leave the sarcoplasmic reticulum. Ca2+ then floods into the sarcoplasm surrounding the actin and myosin filaments (FIGURE 33.7, step 1).

The Ca2+ ions in the sarcoplasm then trigger actin and myosin to interact so that the muscle fiber contracts. How do the Ca2+ ions do this? Recall that strands of the protein tropomyosin are twisted around the chainlike actin polymers (see Figure 33.4), and molecules of the globular protein troponin occur at regular intervals. Each troponin molecule has three subunits: one binds actin, one binds tropomyosin, and one binds Ca2+.
When a muscle fiber is at rest, the tropomyosin strands are positioned so they block the sites on actin filaments where myosin heads can bind. When Ca2+ is released into the sarcoplasm, it binds to troponin, changing troponin’s conformation. Because troponin is bound to tropomyosin, this troponin change affects the conformation of tropomyosin, which twists enough to expose the actin–myosin binding sites (see Figure 33.7, step 2). Myosin heads can then form cross-bridges with actin (step 3). When a myosin head forms a cross-bridge, it is in a conformation that poises it to undergo its “power stroke,” and it is bound to ADP and inorganic phosphate, Pi. Release of Pi activates the power stroke (step 4), during which the myosin head changes conformation, pulling the actin and myosin filaments past each other and releasing ADP. ATP then binds to the myosin head, causing myosin and actin to dissociate (step 5). Hydrolysis of the ATP then occurs, releasing energy (although the ADP and Pi stay bound to myosin) and leading to another conformational change in which myosin returns to its extended conformation (step 6). As long as Ca2+ remains available, the cycle then repeats (step 7).
688
APPLY THE CONCEPT: Muscle cells develop forces by means of cycles of protein–protein interaction
The force that a muscle fiber can generate depends on how many actin–myosin cross-bridges can form. This number in turn depends on the degree of overlap between the actin and myosin filaments in the sarcomeres. This overlap can vary considerably, as shown in Figure 33.3. When a muscle is stretched from its resting length, there is less overlap between the filaments. We can hypothesize that the force a muscle can produce during a given contraction will be a function of its length at the beginning of the contraction relative to its resting length. In an experiment, a bundle of muscle fibers can be attached at either end to an apparatus that can stretch the fibers and measure the force they generate when they are stimulated to contract. The results are shown in the table.
Plot these results with force generated on the y axis, and length of fibers at the beginning of contraction on the x axis. In answering the questions here, keep in mind that “length” refers to a percentage of resting length.
- At what fiber length is a muscle’s force-generating capacity greatest?

- When a muscle is stretched to longer than its resting length, at what percentage of the resting length do you predict that the muscle fibers will not be able to generate any force?
- Why does the force-generating capacity of a muscle decrease at exceptionally great muscle lengths? At exceptionally short lengths?
- When you do a pull-up, which stages of the pull-up are most difficult, and why?
When excitation ends, calcium pumps remove Ca2+ from the sarcoplasm (step 8). The conformations of troponin and tropomyosin then return to their original state, tropomyosin blocks the binding of myosin heads to actin, and contraction stops.
CHECKpoint CONCEPT 33.1
- Imagine planting your feet and trying to push through a concrete wall that’s far too heavy to move. As you push, would you describe the associated muscles in your back, arms, and legs as contracting, shortening, lengthening, or a combination of these words? Explain.
- In a muscle fiber, how is force development aided by the overlapping arrangement of actin and myosin filaments?
- Describe how the concentration of Ca2+ in the sarcoplasmic reticulum of a muscle cell changes before, during, and after the cell is excited by a nerve impulse (action potential).
Now that we have seen how the contractile apparatus of a muscle generates forces, let’s look at how these forces are put to use.