15.2 GENETIC VARIATION AND INDIVIDUAL UNIQUENESS
While examples like sickle-cell anemia, emphysema, and HIV susceptibility show that genetic variation in some genes has important effects on phenotypes, most of the genetic variation in populations is neutral or has no obvious effects. Much of the neutral variation consists of differences in noncoding DNA. Nonetheless, this variation can be revealed by direct studies of DNA that make use of many of the techniques for DNA manipulation described in Chapter 12.
Restriction enzymes, which cleave double-stranded DNA at the positions of specific sequences known as restriction sites, typically four or six nucleotides long, were among the first tools used to study genetic variation in DNA. These enzymes are useful because DNA from different individuals can differ in the distance between adjacent restriction sites or in the presence or absence of a particular restriction site at some location in the genome. Such differences among individuals are the basis of DNA typing, in which the analysis of a small quantity of DNA can uniquely identify an individual. DNA typing can be as reliable as fingerprints for identifying individuals.
15.2.1 Areas of the genome with variable numbers of tandem repeats are useful in DNA typing.
Polymorphisms do not always result from the replacement of one nucleotide or sequence of nucleotides with another. In many cases, as we saw with CCR5, polymorphisms can be differences in the amount of DNA present. When PCR (Chapter 12) is used to amplify a region of the genome from different individuals or from maternal and paternal homologs of the same individual, the lengths of the resulting DNA fragments may be different. They differ because the number of short repeated sequences of DNA varies from one chromosome to the next. This genetic difference is called a variable number tandem repeat, or VNTR (Fig. 15.5). Just as an individual has two copies of each gene, possibly with two different alleles, so each individual has two copies of these polymorphic tandem repeats, each copy representing an allele of the region. The total number of alleles of variable repeats present in a population can be very large. An example with only five alleles is shown in Fig. 15.5a.
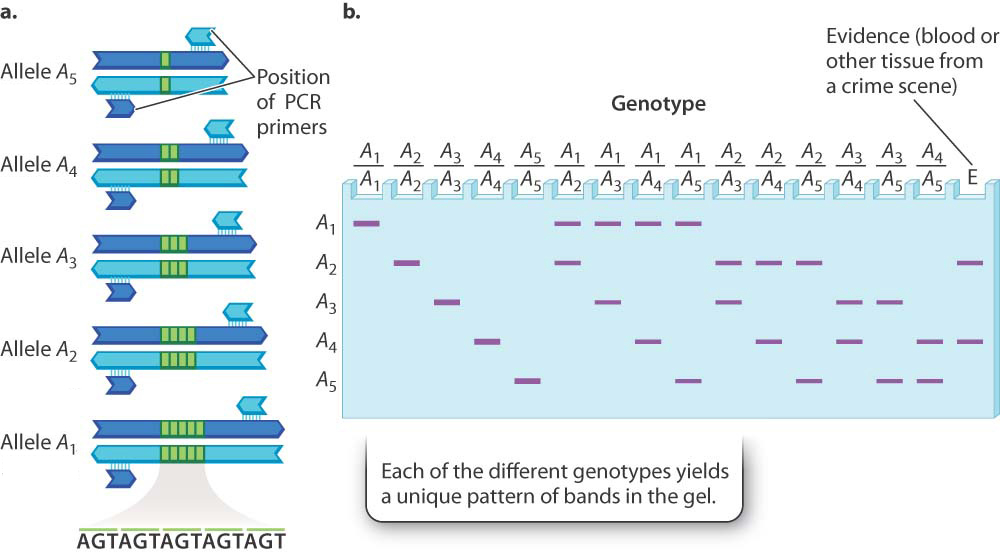
No matter how many alleles exist in the population as a whole, any one diploid individual can have only two alleles, present at the same positions in the homologous chromosomes inherited from the mother and the father. With five alleles there are 5 possible homozygous genotypes and 10 possible heterozygous genotypes. These can be visualized using PCR, in which primers are used to amplify a specific region of DNA and the resulting fragments are separated by size on a gel and then visualized with a dye. The pattern of bands expected from the DNA in each genotype is shown in Fig. 15.5b. Note that each genotype yields a distinct pattern of bands, and so these patterns can be used to identify the genotype of any individual for a particular region of DNA.
With as few as 10 equally frequent alleles, the chance that any two individuals would have the same genotype by chance alone is about 1 in 50, and with 20 equally frequent alleles the chance of identity is about 1 in 200. These numbers suggest why DNA typing can yield a sort of genetic fingerprint that can uniquely match DNA samples.
The lane labeled “E” at the far right in Fig. 15.5b shows the DNA bands for this same polymorphism from biological material collected at the scene of a crime. In this case, the bands in lane E imply that the source of the evidence is an individual of genotype A2/A4 because this is the only genotype with bands with vertical positions in the gel that match those of the evidence sample exactly. A single polymorphism of the type shown in Fig. 15.5 is not enough to say for sure that two pieces of DNA come from the same person, but because the human genome has such polymorphisms scattered throughout, many multiple-allele polymorphisms can be examined. If six or eight of these polymorphisms match between two samples, it is very likely that the samples come from the same individual (or from identical twins). On the other hand, if any of the polymorphisms fails to match band for band, then it is almost certain that the samples come from different individuals (barring such technical mishaps as samples whose DNA is contaminated, degraded, or mislabeled).
One of the advantages of DNA typing is that a large amount of DNA is not needed. Even minuscule amounts of DNA can be typed, so tiny spots of blood, semen, or saliva are sufficient samples. A cotton swab wiped across the inside of your cheek will contain enough DNA for typing; so will a discarded paper cup or a cigarette butt. One enterprising researcher in England used PCR to type the dog feces on his lawn and matched it to the DNA from hair he obtained from neighborhood dogs, much to the chagrin of the guilty dog’s owner.
Quick Check 3
In typing DNA from a sample found at a crime scene, how can a DNA mismatch prove that a suspect is not the source of that sample, whereas a DNA match does not necessarily prove that a suspect is the source?
15.2.2 Some polymorphisms add or remove restriction sites in the DNA.
Researchers can use restriction enzymes to detect another type of polymorphism commonly present in genomes and useful in DNA typing. Sometimes, because of common polymorphisms or sequence differences, a restriction site will be present along a stretch of DNA in some chromosomes but not in others.
Fig. 15.6 shows an example. Here, a restriction enzyme cleaves DNA near the sequence GAGGAG. The three sites shown are in and flanking the β-globin gene from the sickle-cell example we explored earlier in the chapter. The restriction site in the middle includes the GAG that is mutated to GTG in the S allele (see Fig. 15.1). Note that whereas the DNA of the A allele is cleaved at three sites, the DNA of the S allele is cleaved at only two sites because the S mutation changes the GAGGAG restriction site into GTGGAG.
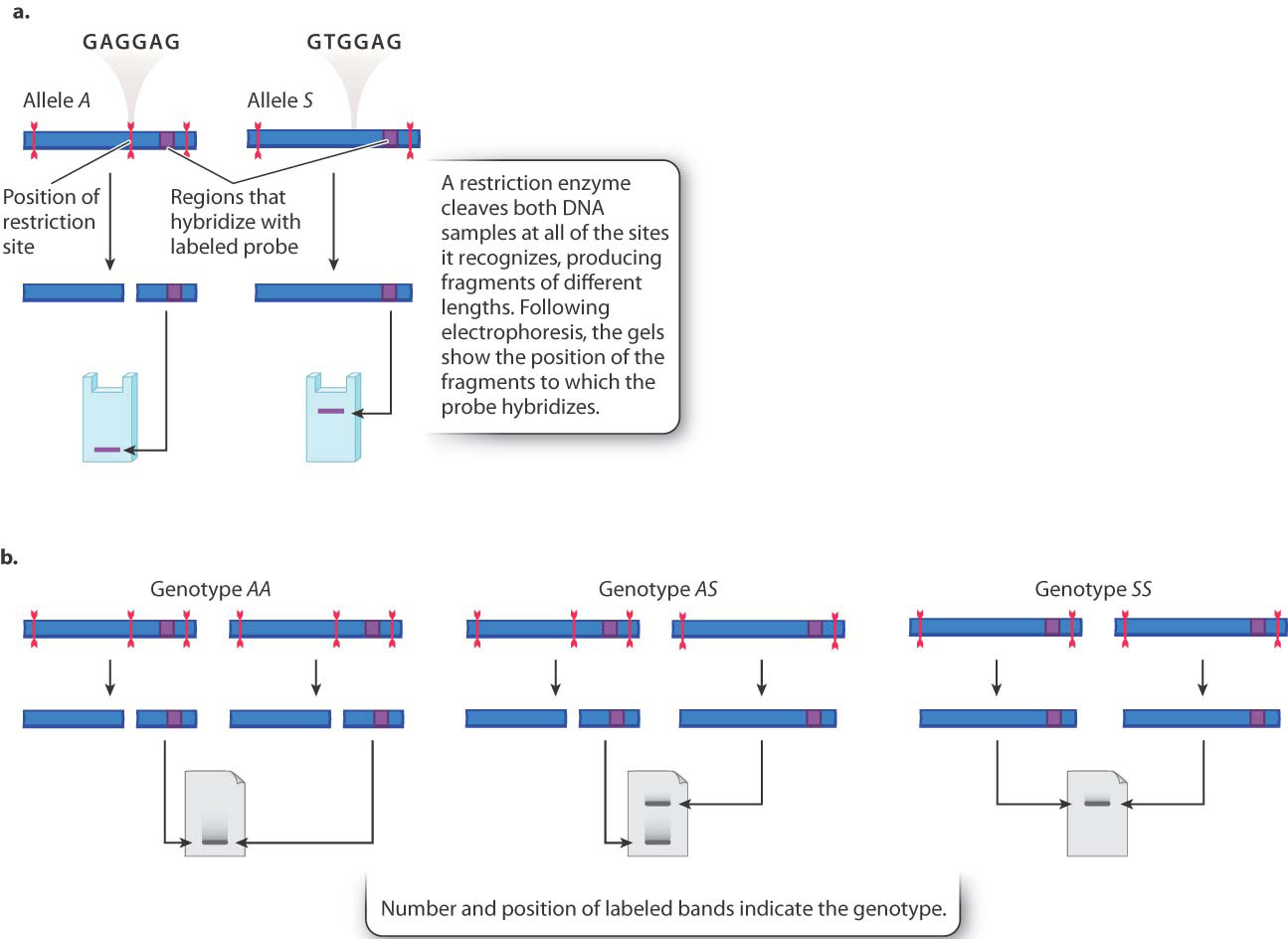
These differences can be visualized by a Southern blot (Chapter 12), in which DNA is isolated from different individuals, cut with restriction enzymes, separated by size on a gel, transferred to a blot, and hybridized to a probe to detect the bands of interest. In this case, we would use a restriction enzyme that recognizes the GAGGAG sequence. The A allele produces two fragments: a short fragment that moves rapidly through the gel and ends up near the bottom, and a longer fragment that ends up near the top. Since the probe hybridizes only where indicated in Fig. 15.6a, the longer restriction fragment produced by digestion of the A allele is not detected. The band produced by the S allele is larger because there is no restriction site in the middle. This larger fragment moves more slowly in gel electrophoresis and ends up forming a band near the top of the gel. This kind of polymorphism is called a restriction fragment length polymorphism, or RFLP, because the length of the restriction fragments is polymorphic—that is, different—in the two alleles.
As seen in the restriction fragment length polymorphism in Fig. 15.6b, DNA from homozygous AA individuals yields only the small fragment, and that from homozygous SS individuals yields only the large fragment. DNA from heterozygous AS individuals yields both the small fragment and the large fragment. Therefore, each of the three genotypes AA, AS, and SS yields a unique pattern of bands, allowing each of the possible genotypes to be identified.
How many restriction fragment length polymorphisms are there in the human genome? As in the case of VNTRs, the human genome is littered with restriction sites, so there are numerous RFLPs in the human population, making it another good individual “fingerprint.”
Quick Check 4
VNTRs and RFLPs both result in restriction fragments of different lengths. How, then, are they different?