20.3 EVOLUTIONARY CONSERVATION OF KEY TRANSCRIPTION FACTORS IN DEVELOPMENT
As we have seen, Hox genes and the proteins they encode are very similar in a wide range of organisms. As a result, they can be identified by their similarity in DNA or amino acid sequence. Molecules that are similar in sequence among distantly related organisms are said to be evolutionarily conserved. Their similarity in sequence suggests that they were present in the most recent common ancestor and have changed very little over time because they carry out a vital function. Many transcription factors important in development are evolutionarily conserved. An impressive example is found in the development of animal eyes.
20.3.1 Animals have evolved a wide variety of eyes.
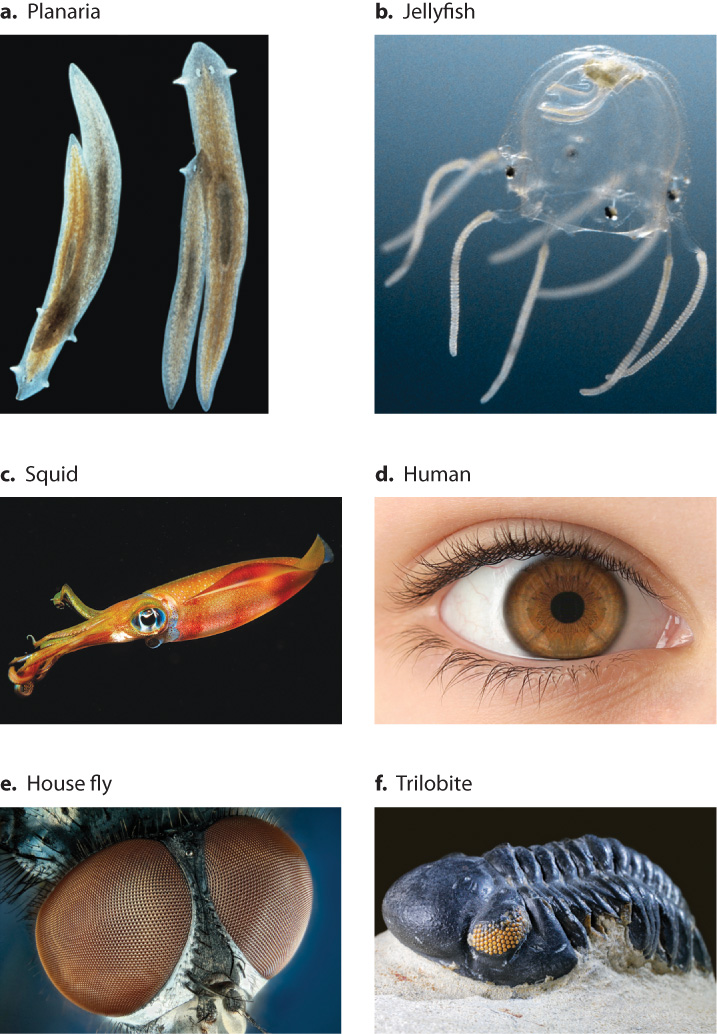
Animal eyes show an amazing diversity in their development and anatomy (Fig. 20.16). Among the simplest eyes are those of the planarian flatworm (Fig. 20.16a), which are only pit-shaped cells containing light-sensitive photoreceptors. The planarian eye has no lens to focus the light, but the animal can perceive differences in light intensity. The incorporation of a spherical lens, as in some jellyfish (Fig. 20.16b), improves the image.
Among the most complex eyes are the camera-type eyes of the squid (Fig. 20.16c) and human (Fig. 20.16d), which have a single lens to focus light onto a light-sensitive tissue, the retina (Chapter 36). Though the single-lens eyes of squids and vertebrates are similar in external appearance, they are vastly different in their development, anatomy, and physiology.
Some organisms, such as the house fly (Fig. 20.16e) and other insects, have compound eyes, that is, eyes consisting of hundreds of small lenses arranged on a convex surface pointing in slightly different directions. This arrangement of lenses allows a wide viewing angle and detection of rapid movement.
In the history of life, eyes are very ancient. By the time of the extraordinary diversification of animals 542 million years ago known as the Cambrian explosion, organisms such as trilobites (Fig. 20.16f) already had well-formed compound eyes. These differed greatly from the eyes in modern insects, however. For example, trilobite eyes had hard mineral lenses composed of calcium carbonate (the world’s first safety glasses, so to speak). Altogether, about 96% of living animal species have true eyes that produce an image, as opposed to simply being able to detect differences in light intensity. Despite this common feature, the extensive diversity among the eyes of different organisms encouraged evolutionary biologists in the belief that the ability to perceive light may have evolved independently about 40 to 60 times.
20.3.2 Pax6 is a master regulator of eye development.
The diversity of animal eyes suggests that they may have evolved independently in different organisms. However, more recent evidence suggests an alternative hypothesis—that they evolved once, very early in evolution and subsequently diverged over time. One argument against the idea of multiple independent origins for light perception is the observation that the light-sensitive molecule in all light-detecting cells is the same, a derivative of vitamin A complexed with a protein called opsin (Chapter 36). The presence of the same light-sensitive molecule in diverse eyes argues that it may have been present in the common ancestor of all animals with eyes and has been retained over time.
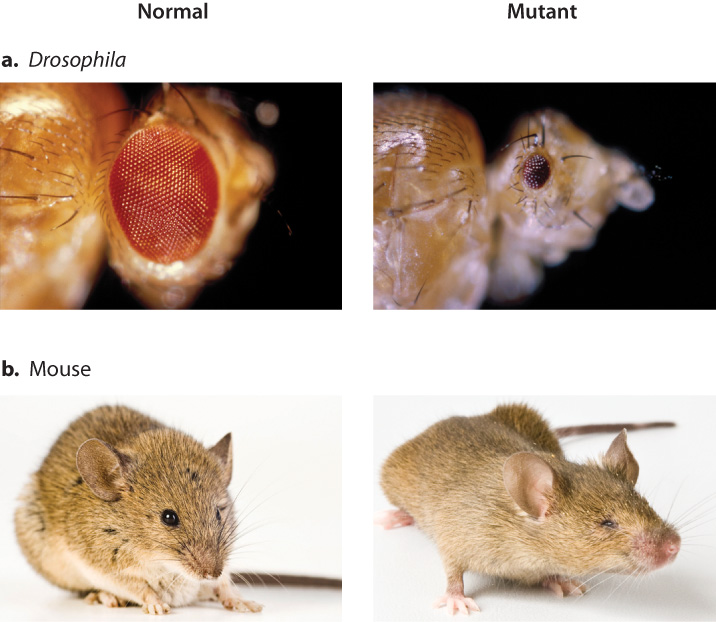
Another argument against multiple independent origins of eyes came from studies of eye development. Researchers identified eyeless, a gene in the fruit fly Drosophila. As its name implies, the phenotype of eyeless mutants is abnormal eye development (Fig. 20.17a). When the protein product of the eyeless gene was identified, it was found to be a transcription factor called Pax6. Mutant forms of a Pax6 gene were already known to cause small eyes in the mouse (Fig. 20.17b) and aniridia (absence of the iris) in humans.
The mutations in the Pax6 gene that cause the development defects in the eye in fruit flies, mice, and humans are loss-of-function mutations. A loss-of-function mutation is just that: a mutation that inactivates the normal function of a gene. In this case, loss-of-function mutations in Pax6 make a defective version of the Pax6 transcription factor that is not able to carry out its function.
The strong conservation of Pax6 in Drosophila and mouse and the similarity in their mutant phenotypes led Swiss developmental biologist Walter Gehring to hypothesize that Pax6 might be a master regulator of eye development. In other words, he hypothesized that Pax6 binds to regulatory regions of a set of genes that turns on a developmental program that induces eye development. In theory, this means that Pax6 can induce the development of an eye in any tissue in which it is expressed.
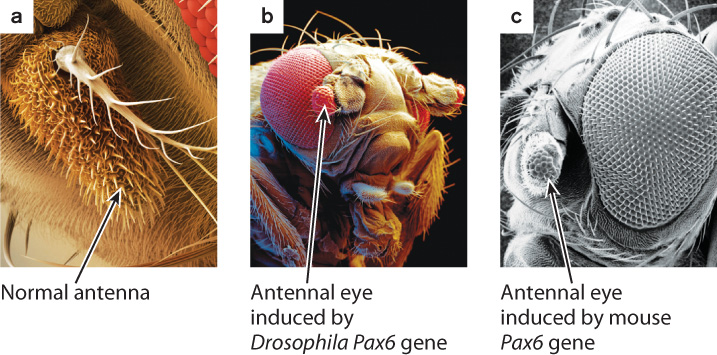
To test this hypothesis, Gehring and collaborators genetically engineered fruit flies that would produce the normal Pax6 transcription factor in the antenna, where it is not normally expressed (Fig. 20.18a). Any mutation in which a gene is expressed in the wrong place or at the wrong time is known as a gain-of-function mutation. (We saw an example of gain-of-function mutations when the Hox genes were expressed in the wrong segments of the fruit fly, producing legs where antennae normally develop and thus resulting in four-winged fruit flies.) For genes that control a developmental pathway, loss-of-function mutations and gain-of-function mutations often have opposite effects on phenotype.
Quick Check 5
For genes that control pathways of development, loss-of-function mutations are usually recessive whereas gain-of-function mutations are usually dominant. Can you suggest a reason why?
Since loss-of-function mutations in Pax6 result in an eyeless phenotype, a gain-of-function mutation should result in eyes developing in whatever tissue Pax6 is expressed. The result is shown in Fig. 20.18b. In the gain-of-function mutation, the antenna developed into a miniature compound eye, and electrical recordings demonstrated that some of these antennal eyes were functional. The researchers also created other gain-of-function mutations that led to eyes on the legs, wings, and other tissues, which the New York Times publicized in an article headlined “With New Fly, Science Outdoes Hollywood.”
Gehring and his group then went one step further. They took the Pax6 gene from mice and expressed it in fruit flies to see whether the mouse Pax6 gene is similar enough to the fruit fly version of the gene that it could induce eye development in the fruit fly. Specifically, they created transgenic fruit flies that expressed the Pax6 gene from mice in the fruit fly antenna. The result is shown in Fig. 20.18c. The mouse gene induced a miniature eye in the fly. Note, however, that the Pax6 gene from mice induced the development of a compound eye of Drosophila, not the single-lens eye of mouse.
The ability of mouse Pax6 to make an eye in fruit flies suggests that mouse and fruit fly Pax6 are not only similar in DNA and amino acid sequence, but also similar in function, and indeed act as a master switch that can turn on a developmental program leading to the formation of an eye. But these observations lead to another question: Why did the mouse Pax6 gene produce fruit fly eyes instead of mouse eyes? The answer is that the fruit fly genome does not include the genes needed to make mouse eyes. Mouse Pax6 protein induces fly eyes because of the downstream genes affected by Pax6, those that function later in the process of eye development. Transcription factors like Pax6 interact with their target genes by binding with short DNA sequences adjacent to the gene, usually at the 5′ end, called cis-regulatory elements. Some transcription factors act as repressors that prevent transcription of the target gene, and others serve as activators by recruiting the transcriptional machinery to the target gene (Chapter 19). A transcription factor can even repress some of its target genes and activate others.
In the fruit fly, Pax6 binds to cis-regulatory elements in many genes, turning some genes on and others off. The products of these downstream genes in turn affect the expression of further downstream genes. The total number of genes that are downstream of Pax6 and that are needed for eye development is estimated at about 2000. Most are not direct targets of Pax6 but are activated indirectly through other transcription factors downstream of Pax6. When mouse Pax6 is expressed in fruit flies, it is similar enough in sequence to activate the genes involved in fruit fly eye development, so it makes sense that mouse Pax6 leads to the development of a fruit fly eye, not a mouse eye.
One scenario of how Pax6 became a master switch for eye development in a wide range of organisms, but produces a diversity of eyes in these organisms, is that Pax6 evolved early in the history of life as a transcription factor able to bind to and regulate genes involved in early eye development. Over time, different genes in different organisms acquired new Pax6-binding cis-regulatory elements by mutation, and if these were beneficial they persisted. The downstream genes that are targets of Pax6 therefore are different in different organisms, but they share two features—they are regulated by Pax6 and they are involved in eye development. So, the early steps are conserved, but the later ones are not.
These studies of Pax6 argue that the genetic pathway for eye development evolved early and is shared among a wide range of animals, even though the eyes that are produced are quite diverse.