20.4 COMBINATORIAL CONTROL IN DEVELOPMENT
Most cis-regulatory elements are located near one or more binding sites for transcription factors, some of which are activators of transcription and others repressors of transcription. The rate of transcription of any gene in any type of cell is therefore determined by the combination of transcription factors that are present in the cell and by the relative balance of activators and repressors. Regulation of gene transcription according to the mix of transcription factors in the cell is known as combinatorial control. Combinatorial control of transcription is another general principle often seen in all multicellular organisms at many stages of development. Here, we discuss flower development as an example of this general principle.
20.4.1 Floral differentiation is a model for plant development.
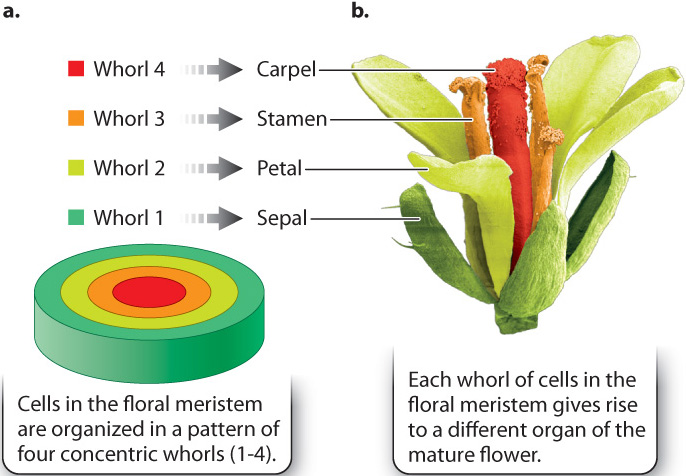
The plant Arabidopsis thaliana, a weed commonly called mouse-ear cress, is a model organism for developmental studies and presents a clear case of combinatorial control. As in all plants, Arabidopsis has regions of undifferentiated cells, called meristems, where growth can take place (Chapter 31). Meristem cells are similar to stem cells in animals. They are the growing points where shoots, roots, and flowers are formed. In floral meristems of Arabidopsis, the flowers develop from a pattern of four concentric circles, or whorls, of cells. From the outermost whorl (whorl 1) of cells to the innermost whorl (whorl 4), the floral organs are formed as shown in Fig. 20.19:
- Cells in whorl 1 form the green sepals, which are modified leaves forming a protective sheath around the petals in the flower bud that unfold like petals when the flower opens.
- Cells in whorl 2 form the petals.
- Cells in whorl 3 form the stamens, the male sexual structures in which pollen is produced. (The small yellow granules in Fig. 20.19b are pollen grains.)
- Cells in whorl 4 form the carpels, the female reproductive structures that receive pollen and contain the ovaries.
20.4.2 The identity of the floral organs is determined by combinatorial control.
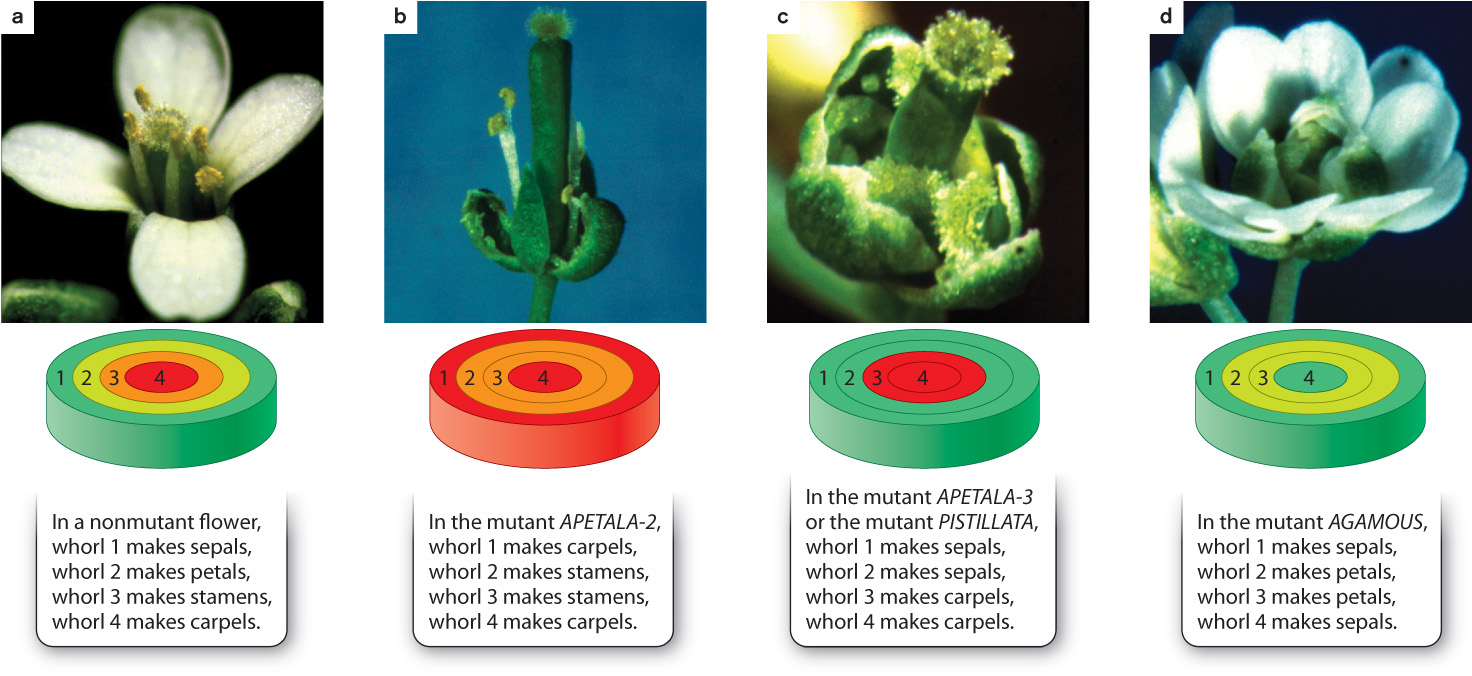
The genetic control of flower development in Arabidopsis was discovered by the analysis of mutant plants, an investigation similar in principle to the way that genetic control of development in fruit flies was determined. The plant mutants fell into three classes showing characteristic floral abnormalities (Fig. 20.20). Mutations in the gene APETALA-2 inactivate the gene’s function and affect whorls 1 and 2 (Fig. 20.20b), those in APETALA-3 or PISTILLATA affect whorls 2 and 3 (Fig. 20.20c), and those in AGAMOUS affect whorls 3 and 4 (Fig. 20.20d). (There are different standards for naming genes and proteins in different organisms. In Arabidopsis, wild-type alleles are written in capital letters with italics, and wild-type proteins in capital letters without italics. Mutant alleles and proteins follow the same rules, but are written in lower-case letters.)
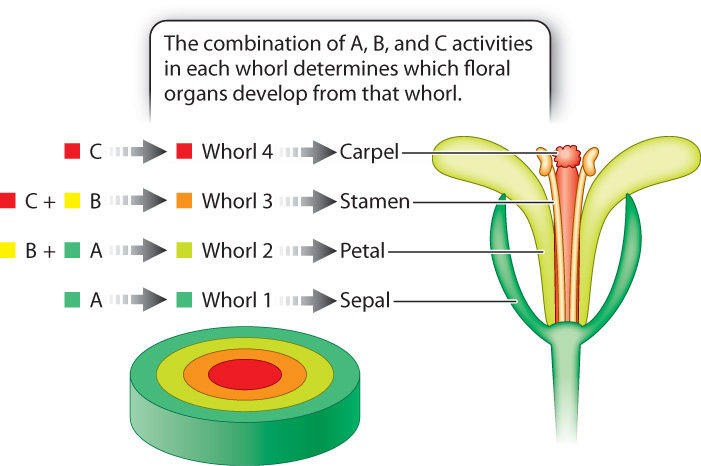
The observation that the mutant phenotypes affect development of pairs of adjacent whorls already hints at combinatorial control, which researchers incorporated into a model of floral development called the ABC model. The model invokes three activities arbitrarily called A, B, and C. These activities represent the function of a protein or proteins that is hypothesized to be present in cells of each whorl as shown in Fig. 20.21. The A activity is present in whorls 1 and 2, B in whorls 2 and 3, and C in whorls 3 and 4. According to the ABC model, activity of A alone results in the formation of sepals, A and B together result in petals, B and C together result in stamens, and C alone results in carpels. Like any good hypothesis (Chapter 1), this one makes specific predictions. Mutants lacking A will have defects in whorls 1 and 2, mutants lacking B will have defects in whorls 2 and 3, and mutants lacking C will have defects in whorls 3 and 4.
Matching these predictions with the mutants in Fig. 20.20 yields the following correspondence:
- The A activity is encoded by the gene APETALA-2.
- The B activity is encoded jointly by the genes APETALA-3 and PISTILLATA.
- The C activity is encoded by the gene AGAMOUS.
To account for the mutant phenotypes in Fig. 20.20, you need only to postulate that when A is absent, C is expressed in whorls 1 and 2 in addition to its normal expression in whorls 3 and 4; and when C is absent, A expression expands into whorls 3 and 4 in addition to its normal expression in whorls 1 and 2. The domains of expression expand in this way because, in addition to its role in activating certain downstream genes, the product of APETALA-2 represses the expression of AGAMOUS in whorls 1 and 2. As a result, AGAMOUS is normally not expressed in whorls 1 and 2. Similarly, in addition to its role in activating other genes, the product of AGAMOUS represses the expression of APETALA-2 in whorls 3 and 4, so APETALA-2 is normally not expressed in whorls 3 and 4.
Quick Check 6
Predict the phenotype of a flower in which APETALA-2 (activity A) and AGAMOUS (activity C) are expressed normally, but APETALA-3 and PISTILLATA (activity B) are expressed in all four whorls.
The identification of the products of these genes demonstrates combinatorial control and explains the mutant phenotypes. All of the genes encode transcription factors, which are called AP2, AP3, PI, and AG, corresponding to the four genes. The transcription factors bind to cis-regulatory elements of genes that encode proteins necessary for each whorl’s development, similar to what we saw earlier for Pax6 and eye development. AP3 and PI are both necessary for the B activity because the proteins form a heterodimer, a protein made up of two different subunits.
Although other transcription factors associated with activities D and E that also contribute to flower development were discovered later, the original ABC model is still valid and serves as an elegant example of combinatorial control. As in the case of the Pax6 gene in the development of animals’ eyes, evolution of the cis-regulatory sequences in downstream genes has resulted in a wide variety of floral morphologies in different plant lineages, some of which are shown in Fig. 20.22.
