Chapter 1. Mirror Experiment Activity for Fig. 6.16: Do pancreatic enzymes function by binding to protein substrates?
Mirror Experiment Activity for Fig. 6.16: Do pancreatic enzymes function by binding to protein substrates?
The experiment described below explored the same concepts as the one described in Figure 6.16 in the textbook. Read the description of the experiment and answer the questions below the description to practice interpreting data and understanding experimental design.
Mirror Experiment activities practice skills described in the brief Experiment and Data Analysis Primers, which can be found by clicking on the “Resources” button on the upper right of your LaunchPad homepage. Certain questions in this activity draw on concepts described in the Data and Data Presentation primer. Click on the “Key Terms” buttons to see definitions of terms used in the question, and click on the “Primer Section” button to pull up a relevant section from the primer.
Experiment
Background
You may know the pancreas as the site of insulin secretion, but this organ also produces enzymes that play a role in digestion. As shown in Fig. 6.16, radiolabeling experiments have demonstrated that enzymes can bind to substrates; however, this work yielded few clues to the physical shape of the enzyme-substrate complexes that were studied. Do pancreatic enzymes also function by binding to protein substrates? If so, can researchers determine the shape of pancreatic enzyme-substrate complexes?
Hypothesis
Edgar Meyer and colleagues hypothesized that pancreatic enzymes are able to form complexes with protein substrates. They also sought to determine the three-dimensional conformation of the enzyme-substrate complexes formed by these enzymes.
Experiment
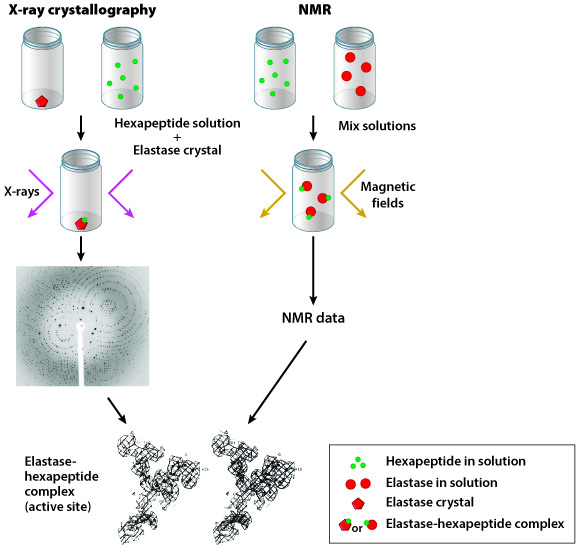
Researchers often use one of two methods to determine the shape of a protein: X-ray crystallography or nuclear magnetic resonance (NMR). As discussed in Fig. 4.5, scientists using X-ray crystallography first produce a crystal of their target protein, then expose this crystal to X-rays. The crystalized protein generates a specific pattern of reflected X-rays, which can be visualized on film (in much the same way as your bones can be visualized in an X-ray). NMR is carried out in a similar manner although the protein does not have to be in crystalline form; however, the protein is exposed to magnetic fields instead of X-rays. In both cases, data collected from X-ray crystallography and NMR experiments help researchers predict the three-dimensional shape of a protein.
Edgar Meyer and colleagues chose to study pig elastase, an enzyme produced by the pancreas; they evaluated the ability of this enzyme to bind to a hexapeptide substrate, a short molecule composed of six amino acids. In their first experiment, an elastase crystal was generated and submerged in a hexapeptide solution; X-ray crystallography was performed on the crystal. In their second experiment, solutions of hexapeptide and uncrystalized elastase were mixed together and NMR was carried out (Figure 1).
Results
Using both X-ray crystallography and NMR techniques, Meyer and colleagues determined that elastase is able to bind a hexapeptide substrate. Furthermore, researchers were able to visualize the three-dimensional structure of this elastase-hexapeptide complex.
Source
Meyer, E. F., Jr., et al., 1988. “Analysis of an enzyme-substrate complex by X-ray crystallography and transferred nuclear Overhauser enhancement measurements: porcine pancreatic elastase and a hexapeptide.” Biochemistry 27: 725-30.
Question
To determine the three-dimensional structure of an enzyme-substrate complex, researchers often pay close attention to X-ray crystallography or NMR results that focus on the active site of an enzyme. Why?
| A. |
| B. |
| C. |
| D. |
| E. |
