Chapter 1. Working With Data 24.8
Working with Data: HOW DO WE KNOW? Fig. 24.8
Fig. 24.8 describes Rebecca Cann’s studies using mitochondrial DNA to map relationships among different populations of humans. Answer the questions after the figure to practice interpreting data and understanding experimental design. Some of these questions refer to concepts that are explained in the following two brief data analysis primers from a set of four available on LaunchPad:
- Experimental Design
- Data and Data Presentation
You can find these primers by clicking on the button labeled “Resources” in the menu at the upper right on your main LaunchPad page. Within the following questions, click on “Primer Section” to read the relevant section from these primers. Click on “Key Terms” to see pop-up definitions of boldfaced terms.
HOW DO WE KNOW?
FIG. 24.8: When and where did the most recent common ancestor of all living humans live?
BACKGROUND With the availability of DNA sequence analysis tools, it became possible to investigate human prehistory by comparing the DNA of living people from different populations.
HYPOTHESIS The multiregional hypothesis of human origins suggests that our most recent common ancestor was living at the time Homo ergaster populations first left Africa, about 2 million years ago.
METHOD Rebecca Cann compared mitochondrial DNA (mtDNA) sequences from 147 people from around the world. This approach required a substantial amount of mtDNA from each individual, which she acquired by collecting placentas from women after childbirth. Instead of sequencing the mtDNA, Cann inferred differences among sequences by digesting the mtDNA with different restriction enzymes, each of which cuts DNA at a specific sequence (Chapter 12). If the sequence is present, the enzyme cuts. If any base in the recognition site of the restriction enzyme has changed in an individual, the enzyme does not cut. The resulting fragments were then separated using gel electrophoresis. By using 12 different enzymes, Cann was able to assay a reasonable proportion of all the mtDNA sequence variation present in the sample.
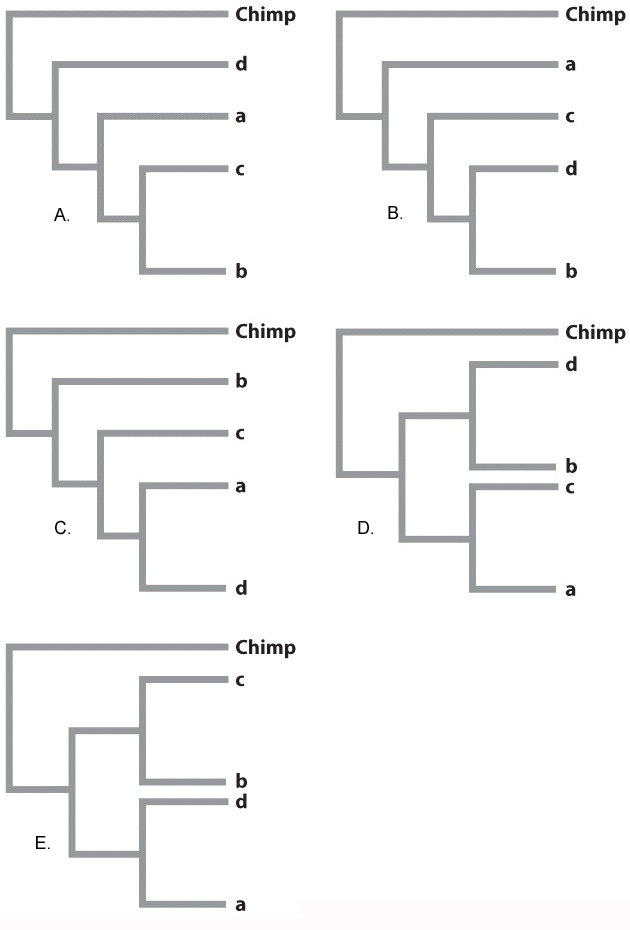
Here, we see a sample dataset for four people and a chimpanzee. There are seven varying restriction sites:
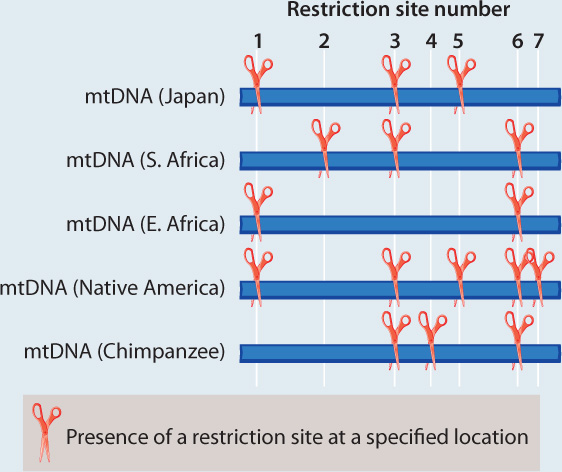
ANALYSIS The data were converted into a family tree by using shared derived characters, described in Chapter 23. By mapping the pattern of changes in this way, the phylogenetic relationships of the mtDNA sequences can be reconstructed. For example, the derived “cut” state at restriction site 1 shared by the East African, Japanese, and Native American sequences implies that the three groups had a common ancestor in which the mutation that created the new restriction site occurred.
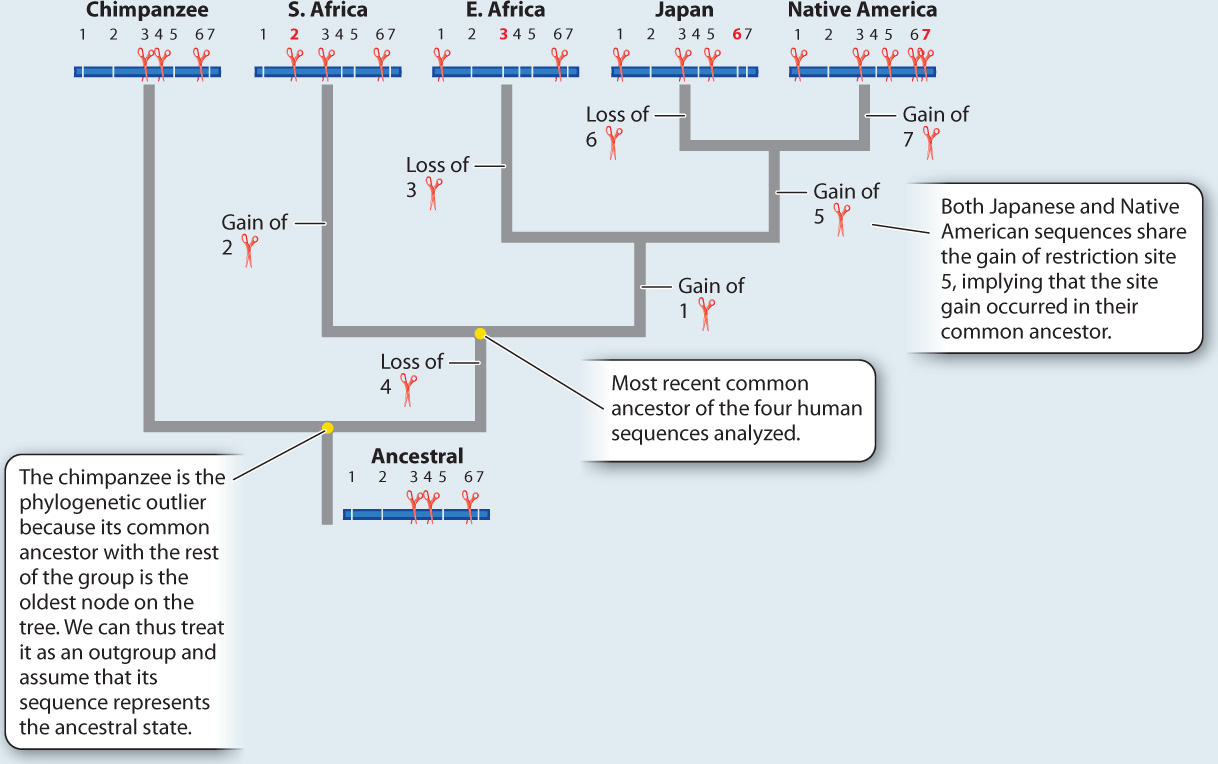
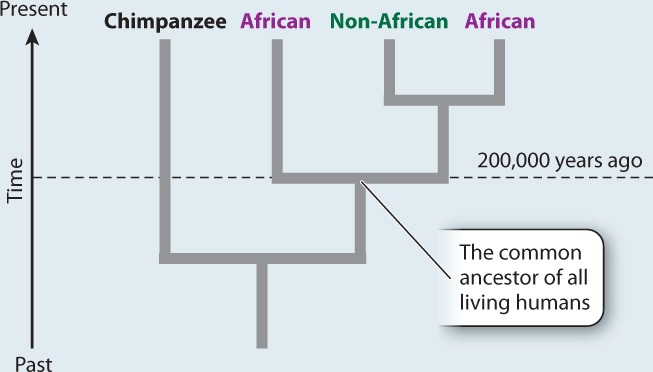
RESULTS AND CONCLUSION A simplified version of Cann’s phylogenetic tree based on mtDNA is shown below. Modern humans arose relatively recently in Africa, and their most common ancestor lived about 200,000 years ago. The multiregional hypothesis is rejected. Also, the data revealed that all non-Africans derive from within the African family tree, implying that H. sapiens left Africa relatively recently. The Cann study gave rise to the out-of-Africa theory of human origins.
FOLLOW-UP WORK This study set the stage for an explosion in genetic studies of human prehistory that used the same approach of comparing sequences in order to identify migration patterns. Today, with molecular tools vastly more powerful than those available to Cann, studies of the distribution of genetic variation are giving us a detailed pattern, even on relatively local scales, of demographic events.
SOURCE Cann, R. L., M. Stoneking, and A. C. Wilson. 1987. “Mitochondrial DNA and Human Evolution.” Nature 325:31–36.
Question
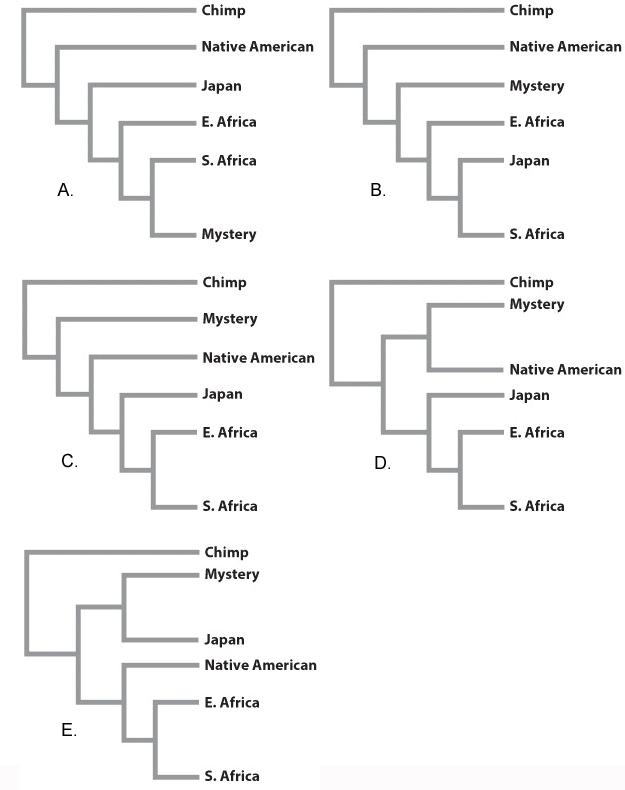
“Found Adrift!” scream the headlines. Aboard a primitive raft in the South Indian Ocean, an extraordinary new life form is found. It’s definitely humanlike, but where does it fit on the human phylogeny?
Dubbed “Mystery,” we add its DNA to our data table in Fig 24.8:
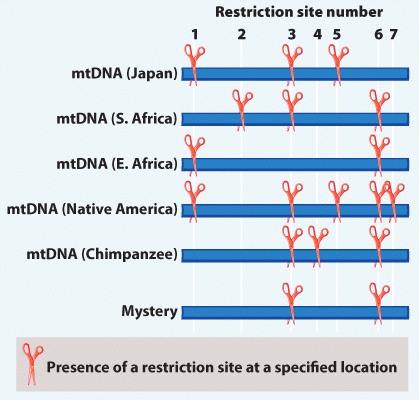
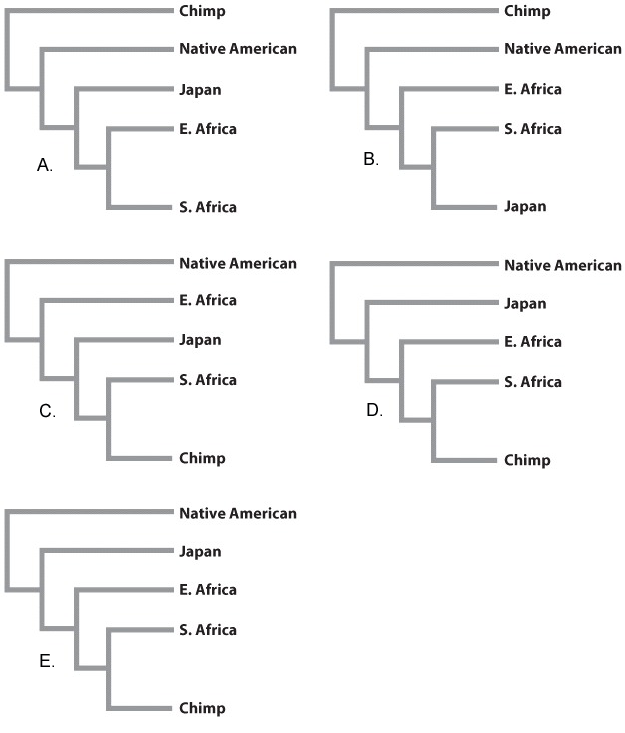
Where should it lie on the phylogeny?
| A. |
| B. |
| C. |
| D. |
| E. |