Producing PCR products with sticky ends
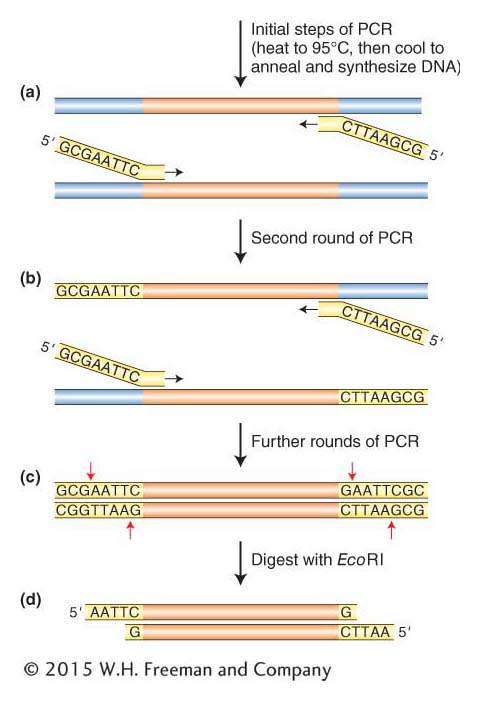
Figure 10-6: Adding EcoRI sites to the ends of PCR products. (a) A pair of PCR primers is designed so that their 3′ ends anneal to the target sequence while their 5′ ends contain sequences encoding the restriction enzyme site (EcoRI in this case). Two additional (random) nucleotides are added to the 5′ end because restriction enzymes require sequences on both sides of the recognition sequence for efficient cutting. The target DNA is denatured, and 5′ ends with the restriction sites remain single stranded while the rest of the primers anneal and are extended by DNA polymerase. (b) In the second round of PCR— only the newly synthesized strands are shown— the DNA primers anneal again, and this time DNA synthesis produces double- stranded DNA molecules just like conventional PCR, but these molecules have restriction sites at one end. (c) The products of the second round and all subsequent rounds have EcoRI sites at both ends. (d) When these are cut with EcoRI, sticky ends are produced.
[Leave] [Close]