Escherichia coli
| Genetic "Vital Statistics" | |
|---|---|
|
Genome size: |
4.6 Mb |
|
Chromosomes: |
1, circular |
|
Number of genes: |
4000 |
|
Percentage with human homologs: |
8% |
|
Average gene size: |
1 kb, no introns |
|
Transposons: |
Strain specific, ~ 60 copies per genome |
|
Genome sequenced in: |
1997 |
Escherichia coli

Key organism for studying:
Transcription, translation, replication, recombination
Mutation
Gene regulation
Recombinant DNA technology
The unicellular bacterium Escherichia coli is widely known as a disease-
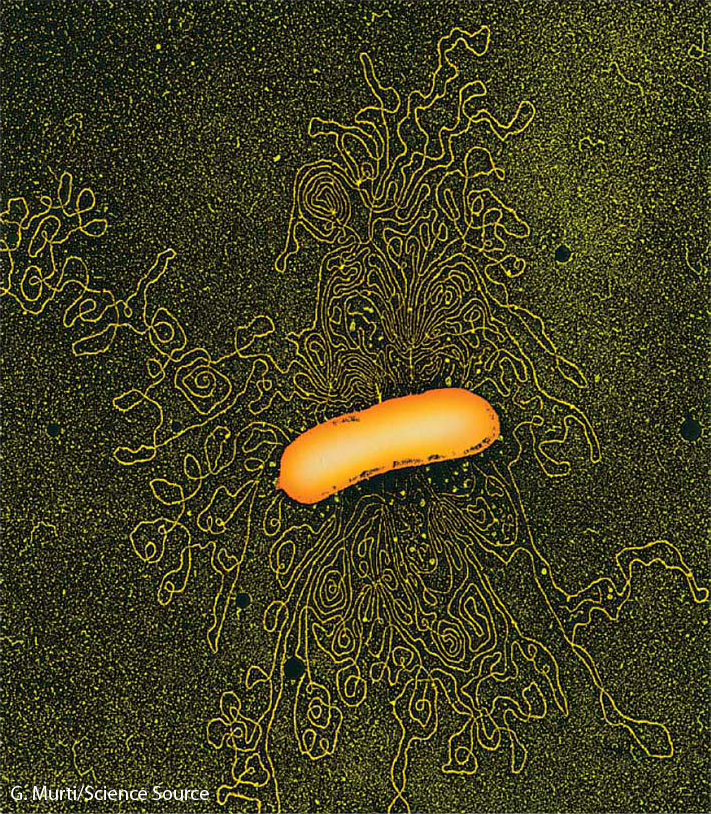
Special features
Much of E. coli’s success as a model organism can be attributed to two statistics: its 1-
Life Cycle
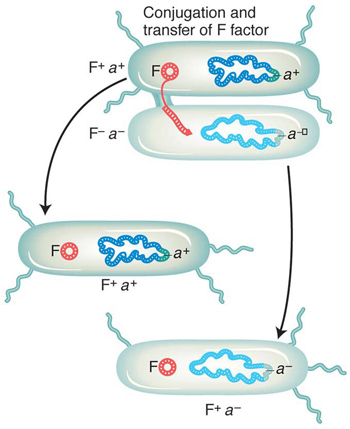
Escherichia coli reproduces asexually by simple cell fission; its haploid genome replicates and partitions with the dividing cell. In the 1940s, Joshua Lederberg and Edward Tatum discovered that E. coli also has a type of sexual cycle in which cells of genetically differentiated “sexes” fuse and exchange some or all of their genomes, sometimes leading to recombination (see Chapter 5). “Males” can convert “females” into males by the transmission of a particular plasmid. This circular extragenomic 100-
Length of life cycle: 20 minutes
795
Geneticists have also taken advantage of some unique genetic elements associated with E. coli. Bacterial plasmids and phages are used as vectors to clone the genes of other organisms within E. coli. Transposable elements from E. coli are harnessed to disrupt genes in cloned eukaryotic DNA. Such bacterial elements are key players in recombinant DNA technology.
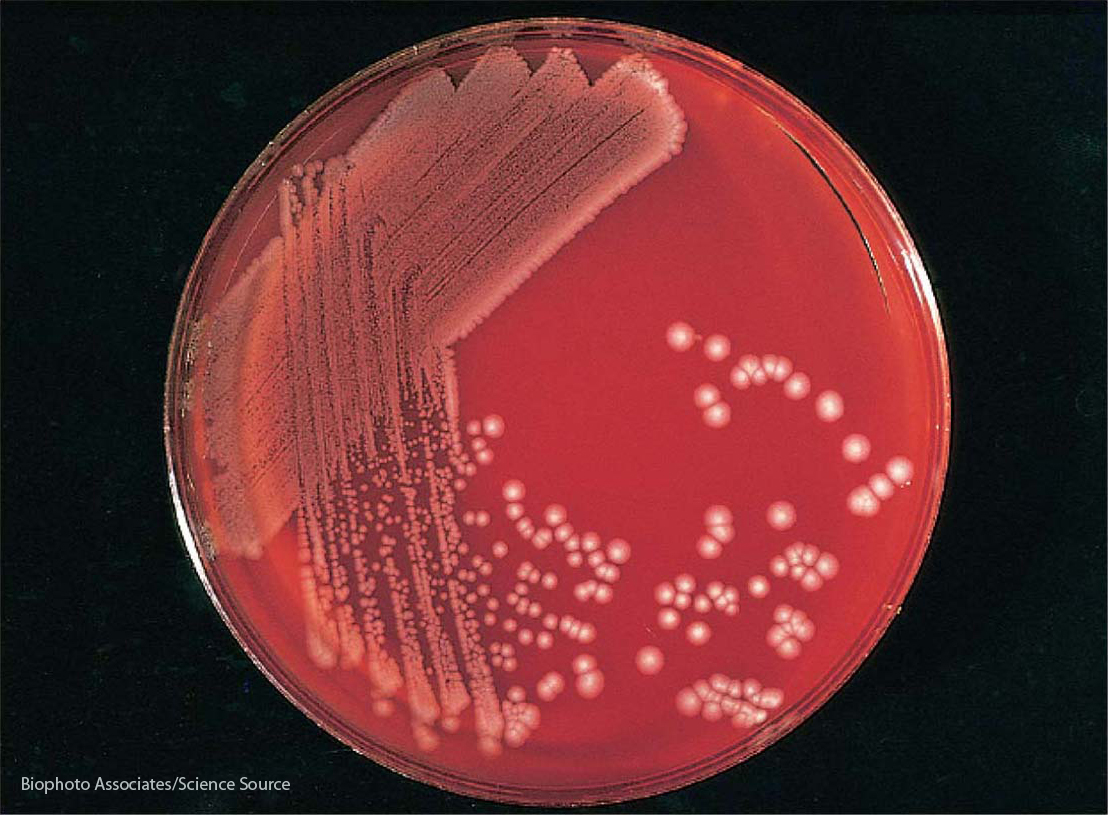
Genetic analysis
Spontaneous E. coli mutants show a variety of DNA changes, ranging from simple base substitutions to the insertion of transposable elements. The study of rare spontaneous mutations in E. coli is feasible because large populations can be screened. However, mutagens also are used to increase mutation frequencies.
To obtain specific mutant phenotypes that might represent defects in a process under study, screens or selections must be designed. For example, nutritional mutations and mutations conferring resistance to drugs or phages can be obtained on plates supplemented with specific chemicals, drugs, or phages. Null mutations of any essential gene will result in no growth; these mutations can be selected by adding penicillin (an antibacterial drug isolated from a fungus), which kills dividing cells but not the nongrowing mutants. For conditional lethal mutations, replica plating can be used: mutated colonies on a master plate are transferred by a felt pad to other plates that are then subjected to some toxic environment. Mutations affecting the expression of a specific gene of interest can be screened by fusing it to a reporter gene such as the lacZ gene, whose protein product can make a blue dye, or the GFP gene, whose product fluoresces when exposed to light of a particular wavelength.
After a set of mutants affecting the process of interest have been obtained, the mutations are sorted into their genes by recombination and complementation. These genes are cloned and sequenced to obtain clues to function. Targeted mutagenesis can be used to tailor mutational changes at specific protein positions.
In E. coli, crosses are used to map mutations and to produce specific cell genotypes (see Chapter 5). Recombinants are made by mixing Hfr cells (having an integrated F plasmid) and F− cells. Generally an Hfr donor transmits part of the bacterial genome, forming a temporary merozygote in which recombination takes place. Hfr crosses can be used to perform mapping by time-
Techniques of Genetic Modification
|
Standard mutagenesis: |
|
|
Chemicals and radiation |
Random somatic mutations |
|
Transposons |
Random somatic insertions |
|
Transgenesis: |
|
|
On plasmid vector |
Free or integrated |
|
On phage vector |
Free or integrated |
|
Transformation |
Integrated |
|
Targeted gene knockouts: |
|
|
Null allele on vector |
Gene replacement by recombination |
|
Engineered allele on vector |
Site- |
Genetic engineering
Transgenesis. E. coli plays a key role in introducing transgenes to other organisms (see Chapter 10). It is the standard organism used for cloning genes of any organism. E. coli plasmids or bacteriophages are used as vectors, carrying the DNA sequence to be cloned. These vectors are introduced into a bacterial cell by transformation, if a plasmid, or by transduction, if a phage, where they replicate in the cytoplasm. Vectors are specially modified to include unique cloning sites that can be cut by a variety of restriction enzymes. Other “shuttle” vectors are designed to move DNA fragments from yeast (“the eukaryotic E. coli”) into E. coli, for its greater ease of genetic manipulation, and then back into yeast for phenotypic assessment.
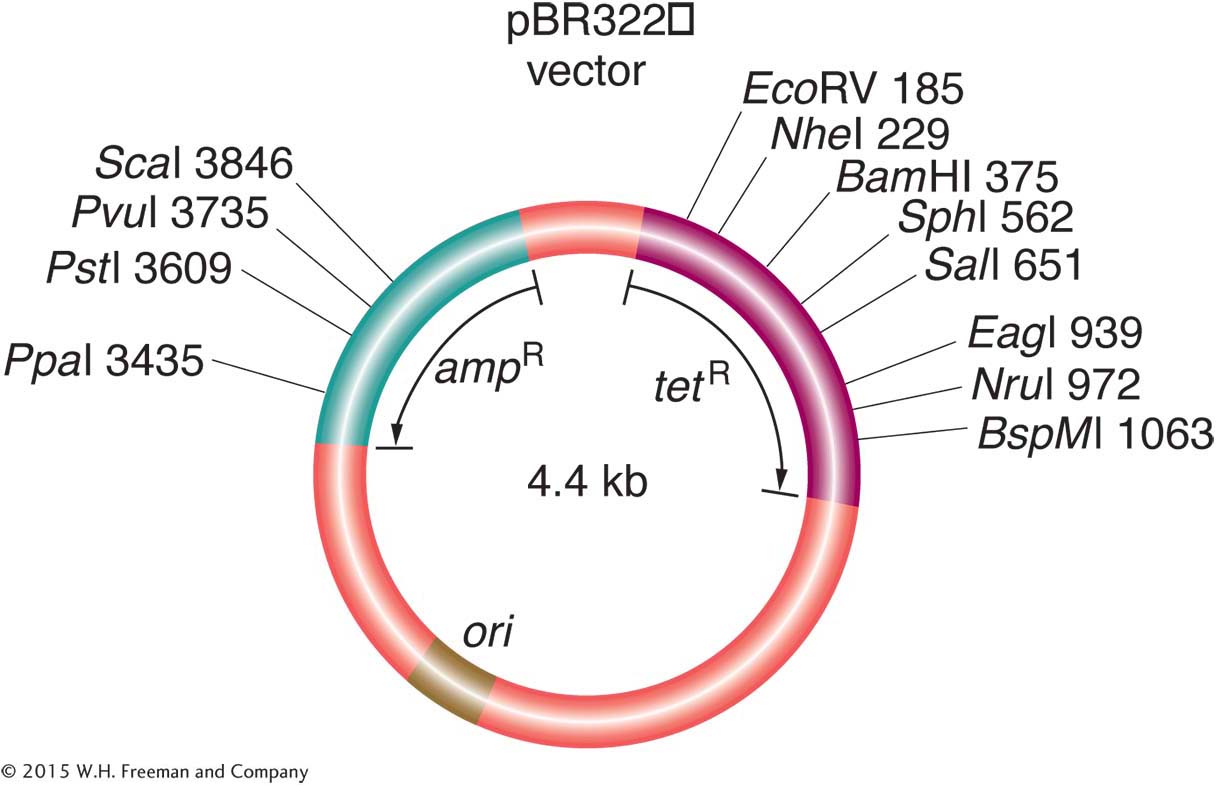
Targeted gene knockouts. A complete set of gene knockouts is being accumulated. In one procedure, a kanamycin-
Main contributions
Pioneering studies for genetics as a whole were carried out in E. coli. Perhaps the greatest triumph was the elucidation of the universal 64-
Other areas of contribution
Cell metabolism
Nonsense suppressors
Colinearity of gene and polypeptide
The operon
Plasmid-
based drug resistance Active transport
796