Chapter 4
13.
|
P |
A d/A d × a D/a D |
|
|
F1 |
A d/a D |
|
|
F2 |
1 A d/A d |
phenotype: A d |
|
|
2 A d/a D |
phenotype: A D |
|
|
1 a D/a D |
phenotype: a D |
16. Because only parental types are recovered, the two genes must be tightly linked and recombination must be very rare. Knowing how many progeny were looked at would give an indication of how close the genes are.
21.
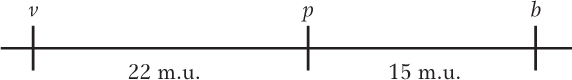
The three genes are linked.
A comparison of the parentals (most frequent) with the double crossovers (least frequent) reveals that the gene order is v p b. There were 2200 recombinants between v and p, and 1500 between p and b. The general formula for map units is
m.u. = 100%(number of recombinants)/total number of progeny
Therefore, the map units between v and p = 100%(2200)/10,000 = 22 m.u., and the map units between p and b = 100%(1500)/10,000 = 15 m.u. The map is
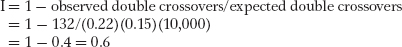
27.
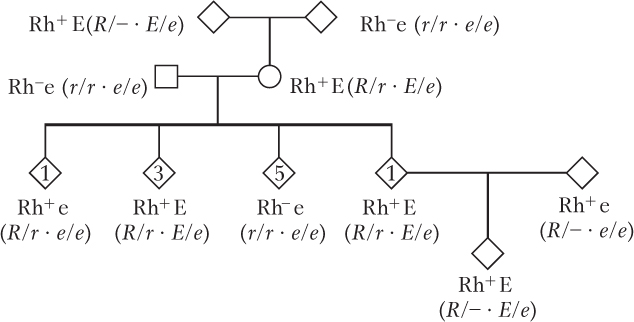
Yes.
Dominant.
As drawn, the pedigree hints at linkage. If unlinked, expect that the phenotypes of the 10 children should be in a 1 : 1 : 1 : 1 ratio of Rh+ E, Rh+ e, Rh− E, and Rh− e. There are actually five Rh− e, four Rh+ E, and one Rh+ e. If linked, this last phenotype would represent a recombinant, and the distance between the two genes would be 100%(1/10) = 10 m.u. However, there is just not enough data to strongly support that conclusion.
33.
a. If the genes are unlinked, the cross is
|
P |
hyg/hyg ; her/her × hyg+/hyg+ ; her+/her+ |
|
F1 |
hyg+/hyg ; her+/her × hyg+/hyg ; her+/her |
|
F2 |
9/16 hyg+/– ; her+/– |
|
|
3/16 hyg+/– ; her/her |
|
|
3/16 hyg/hyg ; her+/– |
|
|
1/16 hyg/hyg ; her/her |
So only 1/16 (or 6.25%) of the seeds are expected to germinate.
b. and c. No. More than twice the expected seeds germinated; so assume that the genes are linked. The cross then is
|
P |
hyg her /hyg her × hyg+her+/hyg+her+ |
|
F1 |
hyg+her+/hyg her × hyg+her+/hyg her |
|
F2 |
13% hyg her/hyg her |
Because this class represents the combination of two parental chromosomes, it is equal to
p(hyg her) × p(hyg her) = (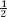 parentals)2 = 0.13
parentals)2 = 0.13
and
parentals = 0.72
So
recombinants = 1 − 0.72 = 0.28
Therefore, a testcross of hyg+ hyg+/hyg her will give
|
36% hyg+her+/hyg her |
|
36% hyg her/hyg her |
|
14% hyg+her/hyg her |
|
14% hyg her+/hyg her |
and 36% of the progeny will grow (the hyg her/hyg her class).
37. The formula for this problem is f (i) = e−mmi/i! where m = 2 and i = 0, 1, or 2.
f (0) = e−220/0! = e−2 = 0.135, or 13.5%
f (1) = e−221/1! = e−2(2) = 0.27, or 27%
f (2) = e−222/2! = e−2(2) = 0.27, or 27%
43.
The cross was pro + × + his, which makes the first tetrad class NPD (6 nonparental ditypes), the second tetrad class T (82 tetratypes), and the third tetrad class PD (112 parental ditypes). When PD >> NPD, you know that the two genes are linked.
Page 835Map distance can be calculated by using the formula RF = [NPD + (1/2)T]100%. In this case, the frequency of NPD is 6/200, or 3%, and the frequency of T is 82/200, or 41%. Map distance between these two loci is therefore 23.5 cM.
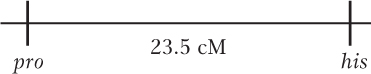
To correct for multiple crossovers, the Perkins formula can be used. Thus, map distance = (T + 6NPD)50%, or (0.41 + 0.18)50% = 29.5 cM.
47.
The cross is W e F/W e F × w E f/w E f and the F1 are W e F/w E f. Progeny that are ww ee ff from a testcross of this F1 must have inherited one of the double-
crossover recombinant chromosomes (w e f). With the assumption of no interference, the expected percentage of double crossovers is 8% × 24% = 1.92%, half of which is 0.96%. To obtain a ww ee ff progeny from a self cross of this F1 requires the independent inheritance of two doubly recombinant w e f chromosomes. Its chances of happening, based on the answer to part a of this problem, are 0.96 × 0.96 = 0.009%.
53. The short answer is that the results tell us little about linkage. Although the number of recombinants (3) is less than the number of parentals (5), one can have no confidence in the fact that the RF is <50%. The main problem is that the sample size is small, so just one individual more or less in a genotypic class can dramatically affect the ratios. Even the chi-
58. The data given for each of the three-
For example, in data set 1, the most common phenotypes (+ + + and a b c) represent the parental-
For data set 2, + b c and a + + (the parentals) should be compared with + + + and a b c (the rarest recombinants) to indicate that the a gene is in the middle. The gene order is b a c.
For data set 3, compare + b + and a + c with a b + and + + c, which gives the gene order b a c.
For data set 4, compare + + c and a b + with + + + and a b c, which gives the gene order a c b.
For data set 5, compare + + + and a b c with + + c and a b +, which gives the gene order a c b.
64.
a. Cross 1 reduces to
|
P |
A/A · B/B · D/D × a/a · b/b · d/d |
|
F1 |
A/a · B/b · D/d × a/a · b/b · d/d |
The testcross progeny indicate that these three genes are linked (CO = crossover, DCO = double crossover).
|
Testcross |
A B D |
316 |
parental |
|
progeny |
a b d |
314 |
parental |
|
|
A B d |
31 |
CO B−D |
|
|
a b D |
39 |
CO B−D |
|
|
A b d |
130 |
CO A−B |
|
|
a B D |
140 |
CO A−B |
|
|
A b D |
17 |
DCO |
|
|
a B d |
13 |
DCO |
A−B: 100%(130 + 140 + 17 + 13)/1000 = 30 m.u.
B−D: 100%(31 + 39 + 17 + 13)/1000 = 10 m.u.
Cross 2 reduces to
|
P |
A/A ·C/C · E/E × a/a · c/c · e/e |
|
F1 |
A/a ·C/c ·E/e × a/a · c/c · e/e |
The testcross progeny indicate that these three genes are linked.
|
Testcross |
A C E |
243 |
parental |
|
progeny |
a c e |
237 |
parental |
|
|
A c e |
62 |
CO A−C |
|
|
a C E |
58 |
CO A−C |
|
|
A C e |
155 |
CO C−E |
|
|
a c E |
165 |
CO C−E |
|
|
a C e |
46 |
DCO |
|
|
A c E |
34 |
DCO |
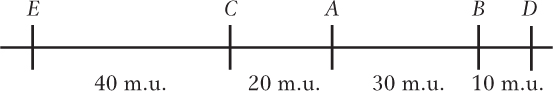
A−B: 100%(62 + 58 + 46 + 34)/1000 = 20 m.u.
B−D: 100%(155 + 165 + 46 + 34)/1000 = 40 m.u.
The map that accommodates all the data is
b Interference (I) = 1 − [(observed DCO)/(expected DCO)]
|
For cross 1: |
I = 1 − {30/[(0.30)(0.10)(1000)]} = 1 − 1 = 0, no interference |
|
For cross 2: |
I = 1 − {80/[(0.20)(0.40)(1000)]} = 1 − 1 = 0, no interference |
69. a. and b. The data support the independent assortment of two genes (call them arg1 and arg2). The cross becomes arg1 ; arg2+ × arg1+ ; arg2 and the resulting tetrads are
|
4 : 0 (PD) |
3 : 1 (T) |
2 : 2 (NPD) |
|
arg1 ; arg2+ |
arg1 ; arg2+ |
arg1 ; arg2 |
|
arg1 ; arg2+ |
arg1+ ; arg2 |
arg1 ; arg2 |
|
arg1+ ; arg2 |
arg1 ; arg2 |
arg1+ ; arg2+ |
|
arg1+ ; arg2 |
arg1+ ; arg2+ |
arg1+ ; arg2+ |
Because PD = NPD, the genes are unlinked.