10.1 Overview: Isolating and Amplifying Specific DNA Fragments
How can a specific segment of DNA be isolated from an entire genome? Furthermore, how can it be isolated in quantities sufficient to analyze features of the DNA such as its nucleotide sequence and its protein product? A crucial insight into the solution to this problem was that researchers could utilize the DNA replication machinery (see Chapter 7) to replicate the DNA segment in question. Such replication is called amplification. It can be done either within live bacterial cells (in vivo) or in a test tube (in vitro).
In the in vivo approach (Figure 10-1a), an investigator begins with a sample of DNA molecules containing the gene of interest. This sample is called the donor DNA, and most often it is an entire genome. Fragments of the donor DNA are inserted into a specially designed plasmid or bacterial virus that will “carry” and amplify the gene of interest and are hence called vectors. First, the donor DNA molecules are cut up by using enzymes called restriction endonucleases as molecular “scissors.” They cut long chromosome-
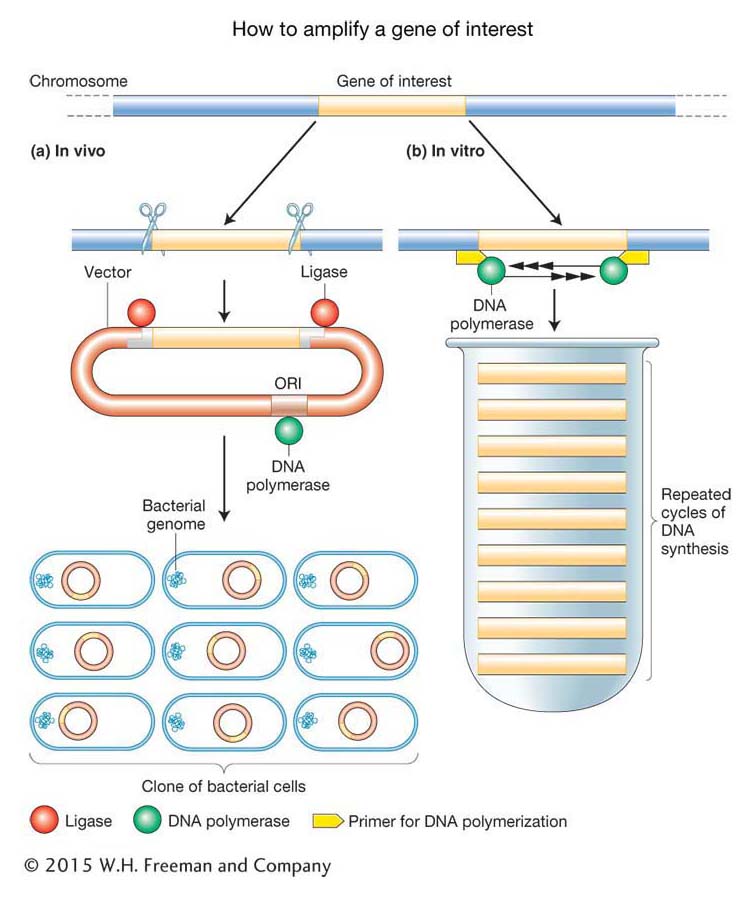
354
In the in vitro approach, called the polymerase chain reaction (PCR) (Figure 10-1b), a specific gene or DNA region of interest is isolated and amplified by DNA polymerase. PCR “finds” the DNA region of interest (called the target DNA) by the complementary binding of specific short synthetic DNA molecules called primers to the ends of that sequence. These primers then guide the replication process directed by the DNA polymerase, which cycles exponentially, resulting in the production of large quantities of the target DNA as an isolated DNA fragment. Even larger quantities of target DNA can be obtained by inserting the PCR product into a plasmid (that is, the in vivo method), thus generating a recombinant DNA molecule like that described above.
We will see repeatedly that DNA technology depends on two basic foundations of molecular biology research:
The ability of specific proteins to recognize and bind to specific base sequences within the DNA double helix (examples are shown in green and red in Figure 10-1).
The ability of complementary single-
stranded DNA or RNA sequences to anneal to form double- stranded molecules (an example is the binding of the primers shown in yellow in Figure 10-1).
The remainder of the chapter will explore some of the uses to which we put amplified DNA. These uses range from routine gene isolation for basic biological research to gene therapy to treat human disease to the production of herbicides and pesticides by crop plants. To illustrate how recombinant DNA is made, let’s consider the cloning of the gene for human insulin, a protein hormone used in the treatment of diabetes. Diabetes is a disease in which blood sugar levels are abnormally high either because the body does not produce enough insulin (type I diabetes) or because cells are unable to respond to insulin (type II diabetes). Mild forms of type I diabetes can be treated by dietary restrictions, but, for many patients, daily insulin treatments are necessary. Until about 30 years ago, cows were the major source of insulin protein. The protein was harvested from the pancreases of animals slaughtered in meatpacking plants and purified on a large scale to eliminate the majority of proteins and other contaminants in the pancreas extracts. Then, in 1982, the first recombinant human insulin came on the market. Human insulin could be made in purer form, at lower cost, and on an industrial scale because it was produced in greater quantities in bacteria by recombinant DNA techniques using the actual human insulin gene sequence. Furthermore, there is no risk of introducing bovine viruses or exciting an immune response against the cows’ insulin. We will use the isolation and production of recombinant insulin as an example of the general steps necessary for making any recombinant DNA.