11.3 Catabolite Repression of the lac Operon: Positive Control
Through a long evolutionary process, the existing lac system has been selected to operate for the optimal energy efficiency of the bacterial cell. Presumably to maximize energy efficiency, two environmental conditions have to be satisfied for the lactose metabolic enzymes to be expressed.
One condition is that lactose must be present in the environment. This condition makes sense because it would be inefficient for the cell to produce the lactose metabolic enzymes if there is no lactose to metabolize. We have already seen that the cell is able to respond to the presence of lactose through the action of a repressor protein.
The other condition is that glucose cannot be present in the cell’s environment. Because the cell can capture more energy from the breakdown of glucose than it can from the breakdown of other sugars, it is more efficient for the cell to metabolize glucose rather than lactose. Thus, mechanisms have evolved that prevent the cell from synthesizing the enzymes for lactose metabolism when both lactose and glucose are present together. The repression of the transcription of lactose-metabolizing genes in the presence of glucose is an example of catabolite repression (glucose is a breakdown product, or a catabolite, of lactose). The transcription of genes encoding proteins necessary for the metabolism of many different sugars is similarly repressed in the presence of glucose. We will see that catabolite repression works through an activator protein.
Page 410
The basics of lac catabolite repression: choosing the best sugar to metabolize
If both lactose and glucose are present, the synthesis of β-galactosidase is not induced until all the glucose has been metabolized. Thus, the cell conserves its energy by metabolizing any existing glucose before going through the energy-expensive process of creating new machinery to metabolize lactose. There are multiple mechanisms that bacteria have evolved to ensure the preferential use of a carbon source and optimal growth. One mechanism is to exclude lactose from the cell. A second mechanism is to regulate operon expression via catabolites.
Figure 11-13: Glucose levels control the lac operon
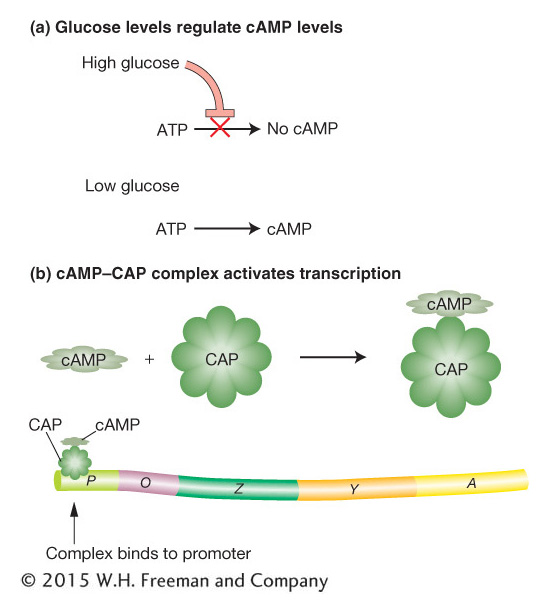
Figure 11-13: Catabolite control of the lac operon. (a) Only under conditions of low glucose is cAMP (cyclic adenosine monophosphate) formed. (b) When cAMP is present, it forms a complex with CAP (catabolite activator protein) that activates transcription by binding to a region within the lac promoter.
The results of studies indicate that the breakdown product of glucose prevents activation of the lac operon by lactose—the catabolite repression just mentioned. The identity of this breakdown product is as yet unknown. However, the glucose breakdown product is known to modulate the level of an important cellular constituent—cyclic adenosine monophosphate (cAMP). When glucose is present in high concentrations, the cell’s cAMP concentration is low. As the glucose concentration decreases, the cell’s concentration of cAMP increases correspondingly (Figure 11-13a). A high concentration of cAMP is necessary for activation of the lac operon. Mutants that cannot convert ATP into cAMP cannot be induced to produce β-galactosidase because the concentration of cAMP is not great enough to activate the lac operon.
What is the role of cAMP in lac activation? A study of a different set of mutants provided an answer. These mutants make cAMP but cannot activate the Lac enzymes because they lack yet another protein, called catabolite activator protein (CAP), encoded by the crp gene. CAP binds to a specific DNA sequence of the lac operon (the CAP-binding site; see Figure 11-14b). The DNA-bound CAP is then able to interact physically with RNA polymerase and increases that enzyme’s affinity for the lac promoter. By itself, CAP cannot bind to the CAP-binding site of the lac operon. However, by binding to cAMP, its allosteric effector, CAP is able to bind to the CAP-binding site and activate transcription by RNA polymerase (Figure 11-13b). By inhibiting CAP when glucose is available, the catabolite-repression system ensures that the lac operon will be activated only when glucose is scarce.
KEY CONCEPT
The lac operon has an added level of control so that the operon is inactive in the presence of glucose even if lactose also is present. An allosteric effector, cAMP, binds to the activator CAP to permit the induction of the lac operon. However, high concentrations of glucose catabolites inhibit production of cAMP, thus failing to produce cAMP–CAP and thereby failing to activate the lac operon.
The structures of target DNA sites
The DNA sequences to which the CAP–cAMP complex binds (see Figure 11-14) are different from the sequences to which the Lac repressor binds. These differences underlie the specificity of DNA binding by these very different regulatory proteins. One property that these sequences do have in common and that is common to many other DNA-binding sites is rotational twofold symmetry. In other words, if we rotate the DNA sequence shown in Figure 11-14 by 180 degrees within the plane of the page, the sequence of the highlighted bases of the binding sites will be identical. The highlighted bases are thought to constitute the important contact sites for protein–DNA interactions. This rotational symmetry corresponds to symmetries within the DNA-binding proteins, many of which are composed of two or four identical subunits. We will consider the structures of some DNA-binding proteins later in the chapter.
Figure 11-14: Many DNA binding sites are symmetrical

Figure 11-14: The DNA base sequences of (a) the lac operator, to which the Lac repressor binds, and (b) the CAP-binding site, to which the CAP–cAMP complex binds. Sequences exhibiting twofold rotational symmetry are indicated by the colored boxes and by a dot at the center point of symmetry.
[(a) Data from W. Gilbert, A. Maxam, and A. Mirzabekov, in N. O. Kjeldgaard and O. Mall0e, eds., Control of Ribosome Synthesis. Academic Press, 1976]
Page 411
How does the binding of the cAMP–CAP complex to the operon further the binding of RNA polymerase to the lac promoter? In Figure 11-15, the DNA is shown as being bent when CAP is bound. This bending of DNA may aid the binding of RNA polymerase to the promoter. There is also evidence that CAP makes direct contact with RNA polymerase that is important for the CAP activation effect. The base sequence shows that CAP and RNA polymerase bind directly adjacent to each other on the lac promoter (Figure 11-16).
Figure 11-15: Binding of CAP DNA
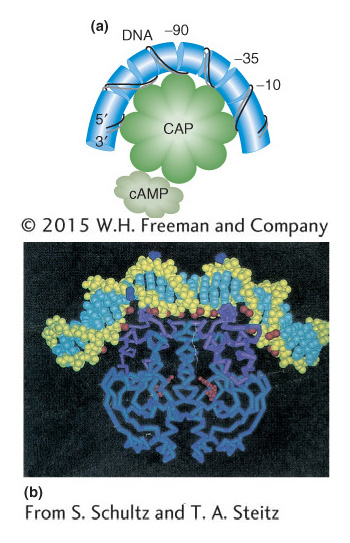
Figure 11-15: (a) When CAP binds the promoter, it creates a bend greater than 90 degrees in the DNA. (b) Image derived from the structural analysis of the CAP-DNA complex.
[(b) From S. Schultz and T. A. Steitz.]
Figure 11-16: CAP and RNA polymerase bind next to each other
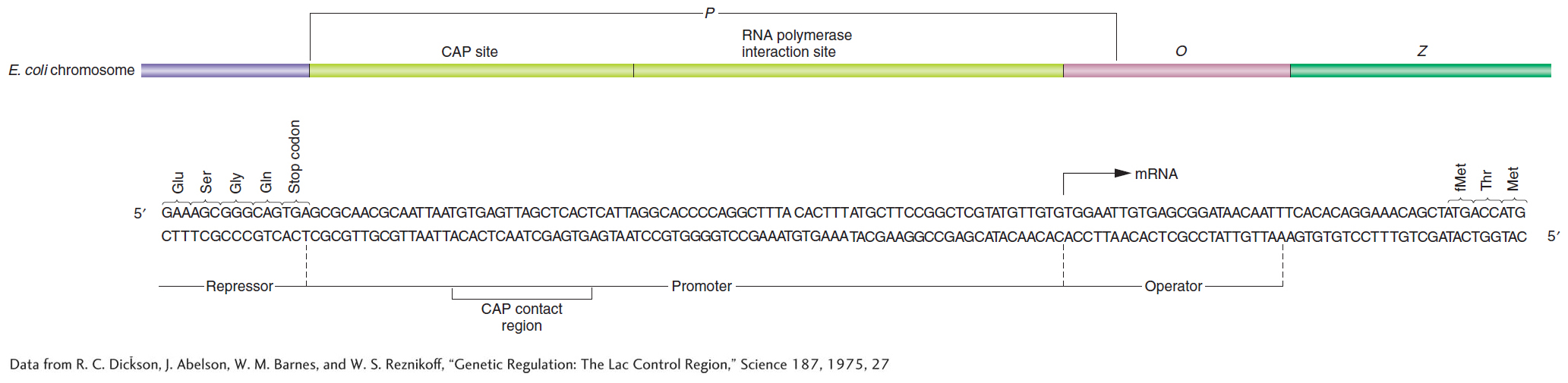
Figure 11-16: The control region of the lac operon. The base sequence and the genetic boundaries of the control region of the lac operon, with partial sequences for the structural genes.
[Data from R. C. Dickson, J. Abelson, W. M. Barnes, and W. S. Reznikoff, “Genetic Regulation: The Lac Control Region,” Science 187, 1975, 27.]
KEY CONCEPT
Generalizing from the lac operon model, we can envision DNA as occupied by regulatory proteins binding to the operator sites that they control. The exact pattern of binding will depend on which genes are turned on or off and whether activators or repressors regulate particular operons.
A summary of the lac operon
We can now fit the CAP–cAMP- and RNA-polymerase-binding sites into the detailed model of the lac operon, as shown in Figure 11-17. The presence of glucose prevents lactose metabolism because a glucose breakdown product inhibits maintenance of the high cAMP levels necessary for formation of the CAP–cAMP complex, which in turn is required for the RNA polymerase to attach at the lac promoter site (see Figure 11-17a,b). Even when there is a shortage of glucose catabolites and CAP–cAMP forms, the mechanism for lactose metabolism will be implemented only if lactose is present (see Figure 11-17c). There are only two or three molecules of β-galactosidase present per cell in the absence of lactose or in the presence of lactose and glucose. This increases to approximately 3000 molecules of enzyme when lactose is present and glucose is absent. Thus, the cell conserves its energy and resources by producing the lactose-metabolizing enzymes only when they are both needed and useful.
Figure 11-17: Negative and positive control of the lac operon
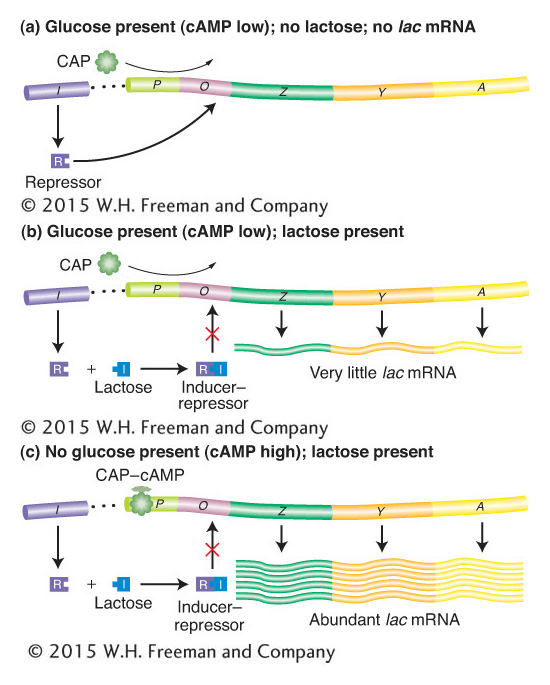
Figure 11-17: The lac operon is controlled jointly by the Lac repressor (negative control) and the catabolite activator protein (CAP; positive control). Large amounts of mRNA are produced only when lactose is present to inactivate the repressor, and low glucose levels promote the formation of the CAP–cAMP complex, which positively regulates transcription.
Page 412
Inducer-repressor control of the lac operon is an example of repression, or negative control, in which expression is normally blocked. In contrast, the CAP–cAMP system is an example of activation, or positive control, because it acts as a signal that activates expression—in this case, the activating signal is the interaction of the CAP–cAMP complex with the CAP-binding site on DNA. Figure 11-18 outlines these two basic types of control systems.
Figure 11-18: Repression and activation compared
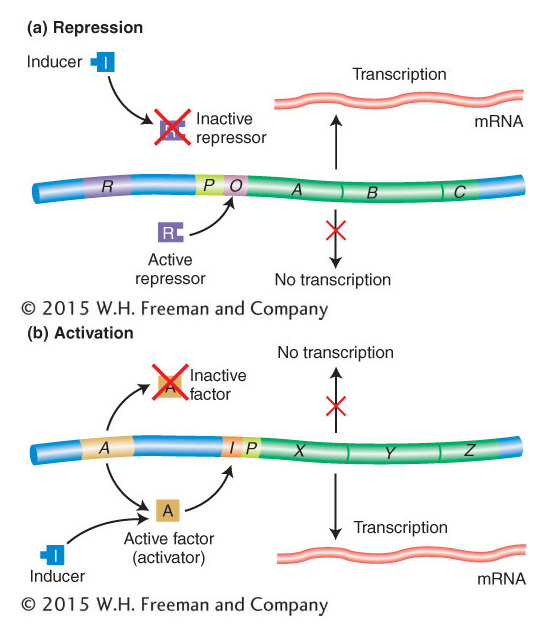
Figure 11-18: (a) In repression, an active repressor (encoded by the R gene in this example) blocks expression of the A, B, C operon by binding to an operator site (O). (b) In activation, a functional activator is required for gene expression. A nonfunctional activator results in no expression of genes X, Y, Z. Small molecules can convert a nonfunctional activator into a functional one that then binds to the control region of the operon, termed I in this case. The positions of both O and I with respect to the promoter P in the two examples are arbitrarily drawn, inasmuch as their positions differ in different operons.
Page 413
KEY CONCEPT
The lac operon is a cluster of structural genes that specify enzymes taking part in lactose metabolism. These genes are controlled by the coordinated actions of cis-acting promoter and operator regions. The activity of these regions is, in turn, determined by repressor and activator molecules specified by separate regulator genes.





