11.7 Alternative Sigma Factors Regulate Large Sets of Genes
Thus far, we have seen how single switches can control the expression of single operons or two operons containing as many as a couple of dozen genes. Some physiological responses to changes in the environment require the coordinated expression of large sets of unlinked genes located throughout the genome to bring about dramatic physiological and even morphological changes. Analyses of these processes have revealed another twist in bacterial gene regulation: the control of large numbers of genes by alternative sigma (σ) factors of RNA polymerase. One such example, the process of sporulation in Bacillus subtilis, has been analyzed in great detail in the past few decades. Under stress, the bacterium forms spores that are remarkably resistant to heat and desiccation.
Early in the process of sporulation, the bacterium divides asymmetrically, generating two components of unequal size that have very different fates. The smaller compartment, the forespore, develops into the spore. The larger compartment, the mother cell, nurtures the developing spore and lyses when spore morphogenesis is complete to liberate the spore (Figure 11-31a). Genetic dissection of this process has entailed the isolation of many mutants that cannot sporulate. Detailed investigations have led to the characterization of several key regulatory proteins that directly regulate programs of gene expression that are specific to either the forespore or the mother cell. Four of these proteins are alternative σ factors.
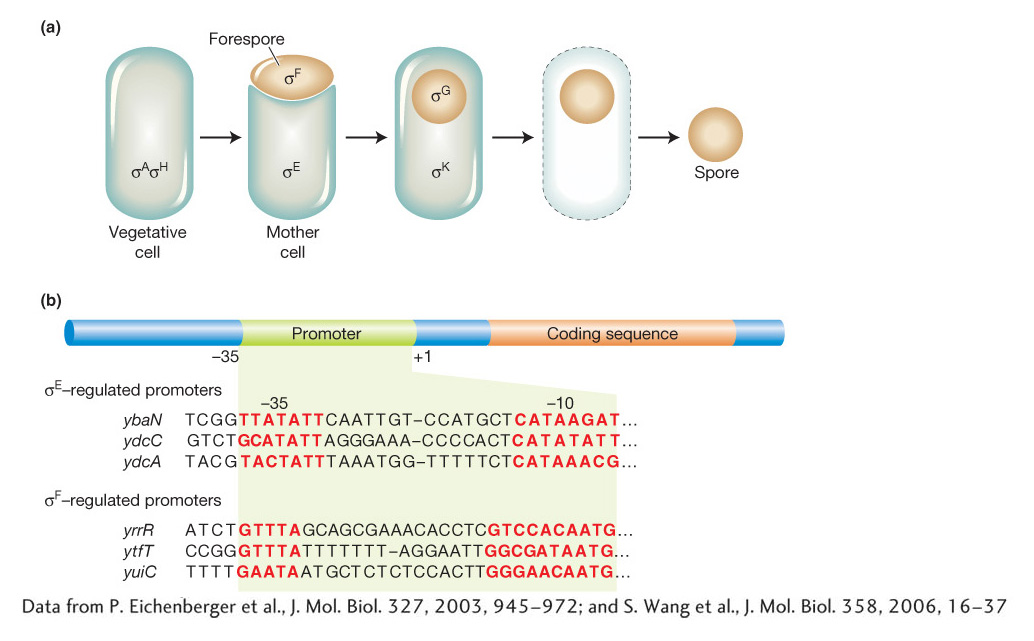
Recall that transcription initiation in bacteria includes the binding of the σ subunit of RNA polymerase to the −35 and −11 regions of gene promoters. The σ factor disassociates from the complex when transcription begins and is recycled. In B. subtilis, two σ factors, σA and σH, are active in vegetative cells. During sporulation, a different σ factor, σF, becomes active in the forespore and activates a group of more than 40 genes. One gene activated by σF is a secreted protein that in turn triggers the proteolytic processing of the inactive precursor pro-
How do these alternative σ factors control different aspects of the sporulation process? The answers have become crystal clear with the advent of new approaches for characterizing the expression of all genes in a genome (see Section 14.6). It is now possible to monitor the transcription of each B. subtilis gene during vegetative growth and spore formation and in different compartments of the spore. Several hundred genes have been identified in this fashion that are transcriptionally activated or repressed during spore formation.
How are the different sets of genes controlled by each σ factor? Each σ factor has different sequence-
KEY CONCEPT
Sequential expression of alternative σ factors that recognize alternative promoter sequences provides for the coordinated expression of large numbers of independent operons and unlinked genes during the developmental program of sporulation.Alternative σ factors also play important roles in the virulence of human pathogens. For example, bacteria of the genus Clostridium produce potent toxins that are responsible for severe diseases such as botulism, tetanus, and gangrene. Key toxin genes of C. botulinum, C. tetani, and C. perfringens have recently been discovered to be controlled by related, alternative σ factors that recognize similar sequences in the −35 and −10 regions of the toxin genes. Understanding the mechanisms of toxin-