SOLVED PROBLEMS
SOLVED PROBLEM 1. About 70 percent of all Caucasians can taste the chemical phenylthiocarbamide, and the remainder cannot. The ability to taste this chemical is determined by the dominant allele T, and the inability to taste is determined by the recessive allele t. If the population is assumed to be in Hardy–
Solution
Because 70 percent are tasters (T/T and T/t), 30 percent must be nontasters (t/t). This homozygous recessive frequency is equal to q2; so, to obtain q, we simply take the square root of 0.30:
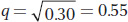
Because p + q = 1, we can write p = 1 − q = 1 − 0.55 = 0.45.
Now we can calculate
|
p2 = (0.45)2 = 0.20, |
the frequency of T/T |
|
2pq = 2 × 0.45 × 0.55 = 0.50, |
the frequency of T/t |
|
q2 = 0.3, |
the frequency of t/t |
SOLVED PROBLEM 2. In a large experimental Drosophila population, the relative fitness of a recessive phenotype is calculated to be 0.90, and the mutation rate to the recessive allele is 5 × 10−5. If the population is allowed to come to equilibrium, what allele frequencies can be predicted?
Solution
Here, mutation and selection are working in opposite directions, and so an equilibrium is predicted. Such an equilibrium is described by the formula
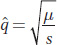
In the present question,
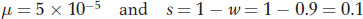
Hence,
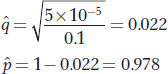
SOLVED PROBLEM 3. A colony of 50 horned puffins (Fratercula corniculata) is established at a zoo and maintained there for 30 generations.
a. If the inbreeding coefficient of the founding members was zero (F = 0.0), what is the expected inbreeding coefficient for this population at present?
b. For a deleterious disease allele with a frequency of 0.001 in the wild, what is the predicted frequency of homozygous affected birds in the wild and in the zoo population at present?
Solution
a. In Box 18-
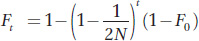
Substituting in N = 50, t = 30, and F0 = 0, we obtain
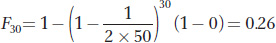
b. If the frequency of a recessive disease allele (q) in the wild is 0.001, then by applying the Hardy–
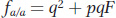
Substituting in q = 0.001, p = 0.999, and F = 0.26, we obtain
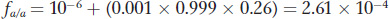
The ratio of 2.61 × 10−4 to 10−6 shows us that there is a 261-
SOLVED PROBLEM 4. At a criminal trial, the prosecutor presents genotypes for three microsatellite loci from the FBI CODIS set. He reports that a DNA sample from the crime scene and one from the suspect both have the genotype FGA1/FGA4, TPOX1/TPOX3, VWA2/VWA7 at these three microsatellites. He also presents the allelic frequencies for the general population to which the suspect belongs (see the table that follows). What is the probability that the genotype of the DNA evidence would match that of the suspect given that the person who committed the crime and the suspect are different individuals? What assumptions do you make when calculating this probability?
|
Allele |
Frequency |
|---|---|
|
FGA1 |
0.30 |
|
FGA4 |
0.26 |
|
TPOX1 |
0.32 |
|
TPOX3 |
0.65 |
|
VWA2 |
0.23 |
|
VWA7 |
0.59 |
Solution
The probability that the genotype of the DNA evidence matches that of the suspect given that the person who committed the crime and the suspect are different individuals is the same as the probability that a randomly chosen member of the population would have the same genotype as the DNA evidence. The probability of a randomly chosen person being FGA1/FGA4 = 2pq = 2(0.30) (0.26) = 0.156 and, similarly, the probability of a random person being TPOX1/TPOX3 = 0.416 and VWA2/VWA7 = 0.2714. Applying the multiplicative rule, the probability of a random member of the population being FGA1/FGA4, TPOX1/TPOX3, VWA2/VWA7 = 0.156 × 0.416 × 0.2714 = 0.0176. In calculating this probability, we have assumed that the population is at Hardy–