Chapter 2. Finding genes for common diseases
Introduction
Finding genes for common diseases
Introduction
In 2007, a consortium of fifty British research groups collectively known as the Wellcome Trust Case Control Consortium (WTCCC) investigated the genetic basis of seven common human diseases—
1. Genome-
Basis of the Global Association Study
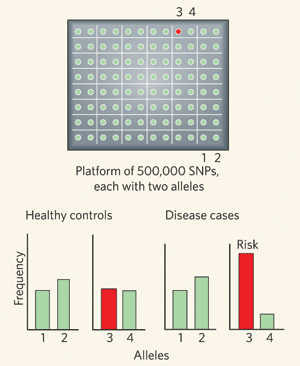
Figure 1 Source: Figure 1 from A. M. Bowcock. Genomics: Guilt by association. Nature 447, 645-
In one GWAS, the WTCCC investigators examined genetic variation at 500,000 different positions within the genomes of 17,000 individuals living in the United Kingdom (Figure 1). This statistical approach compares the frequencies of genetic variation in individuals with a given disease and in control individuals from the same population. There are approximately ten million such changes (known as single nucleotide polymorphisms, or SNPs) located at particular positions scattered throughout the human genome where variation is found in at least 10% of the population. In a GWAS, researchers examine the relationship between every DNA position and a particular trait (such as diabetes) using the signal from each position as an indicator for the DNA sequence that surrounds it. A strong “association'” between a DNA position and a particular disease or trait marks the general location of the associated genetic alteration, even if the associated SNP itself is not directly responsible for the disease (discussed in your textbook in Section 19.6).
The concept of drawing an association between biological traits and disease is not new2, but the overall scope and scale of the WTCCC in its application of this concept was unparalleled. The following factors were crucial to the success of this study and to keeping costs within reasonable limits: access to DNA samples from large numbers of unrelated patients from the United Kingdom; knowledge of the complete sequence of the human genome; the availability of a vast number of SNPs3; the completed HapMap project4, which provided information about the genetic relatedness of SNPs; and the availability of high-
2. Billington, B. P. Gastric cancer; relationships between ABO blood-
3. Carlson, C. S. et al. Selecting a maximally informative set of single-
4. A haplotype map of the human genome. Nature 437, 1299–
WTCCC Findings and Follow-up Studies
Among the seven common diseases, the WTCCC study yielded statistically significant evidence for genetic associations with twelve previously identified genomic regions and with twelve new genomic regions (Figure 2). Although the WTCCC report provided an initial glimpse of these associations, independent studies by other research groups subsequently confirmed all but one of the most significant regions identified by the WTCCC through replication studies5, 6, 7. Furthermore, follow-
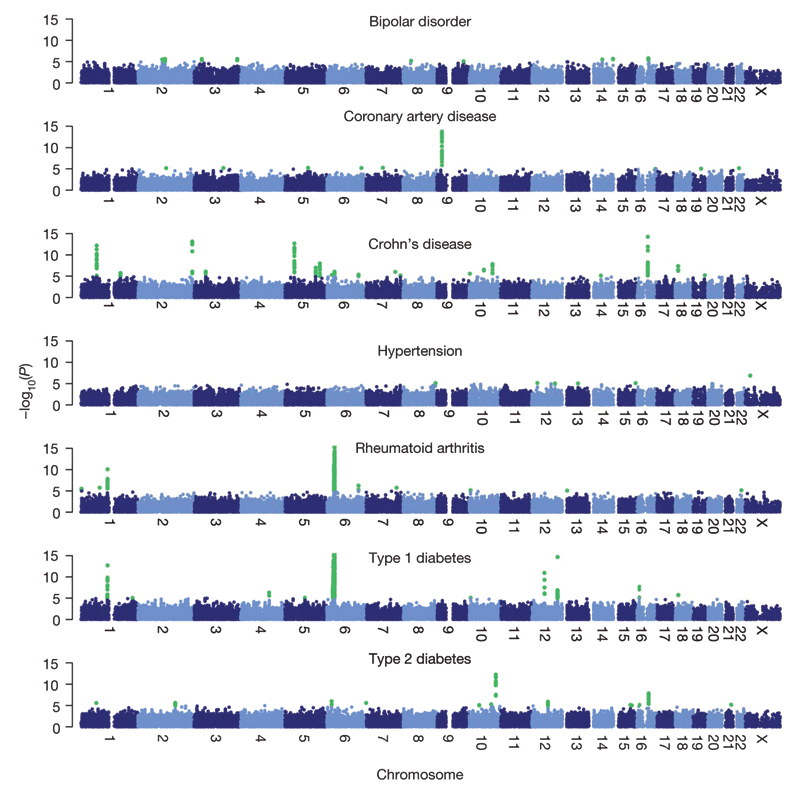
Figure 4 from Wellcome Trust Case Control Consortium. Genome-
The WTCCC data are publicly available and have proven to be an invaluable resource to other groups and consortia engaged in similar studies to identify genetic markers linked to these and other diseases. Even larger studies have been performed since the WTCCC study, in some cases involving more than 100,000 individuals9, 10. One is a study of body mass with approximately 2.8 million SNPs, revealing thirty-
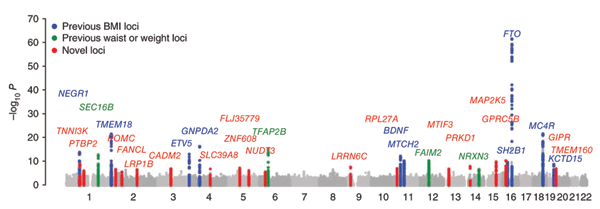
Figure 1 (panel a only) from E. K. Speliotes et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics 42, 937-
5. Todd, J. A. et al. Robust associations of four new chromosome regions from genome-
6. Zeggini, E. et al. Replication of genome-
7. Saxena, R. et al. Genome-
8. D'Addabbo, A. et al. Discovering genetic variants in Crohn's disease by exploring genomic regions enriched of weak association signals. Digestive and Liver Disease [Epub ahead of print] (2011). doi: 10.1016/j.dld.2011.02.010
9. Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707-
10. Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832-
11. Speliotes, E. K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics 42, 937-
12. Manolio, T. A. Genomewide association studies and assessment of the risk of disease. New England Journal of Medicine 363, 166-
The Next Step Following a GWAS
After a particular disease or trait has been associated with a particular marker SNP, what happens? The next step is to move beyond the associated marker SNP to study the exact nature of the variant that is responsible for the disease or trait. At what types of locations within the genome do the responsible variants typically occur? Overall, it seems that the variants leading to common diseases are diverse: only approximately 10% are located within genes where they alter coding sequences13, whereas 45% are located within the noncoding sequences of genes and approximately 45% are located between genes; some variants even lie within gene deserts14, which are chromosomal regions that contain few or no genes. The approximately 90% of variants that are located outside of gene coding sequences are expected to be involved in gene regulation, but identifying the true variants and understanding their biological role remain formidable challenges.
13. Hindorff, L. A. et al. Potential etiologic and functional implications of genome-
14. Libioulle, C. et al. Novel Crohn disease locus identified by genome-
The First GWAS
The first study demonstrating that a GWAS might be a feasible approach for identifying genes responsible for a common disease was focused on age-
15. Klein, R. J. et al. Complement factor H polymorphism in age-
16. Sobrin, L. et al. Genetic profile for five common variants associated with age-
Crohn's Disease
In the WTCCC study, one of the newly identified susceptibility genes for the inflammatory bowel disease Crohn's disease was autophagy-
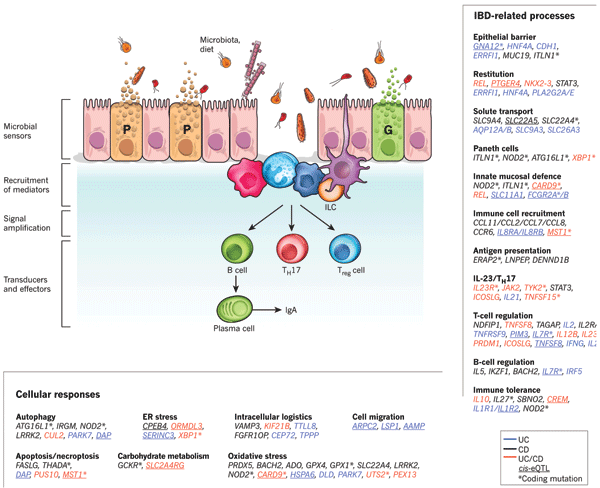
Figure 2 from Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307-
17. Budarf, M. L. et al. GWA studies: Rewriting the story of IBD. Trends in Genetics 25, 137-
18. Franke, A. et al. Genome-
Benefits of GWAS
Why are GWAS approaches so valuable? A GWAS can implicate previously unsuspected pathways that are altered during disease, which can lead to the development of new therapies. For example, the discovery that inflammation plays a role in AMD is now being used to develop novel forms of treatment. In addition, knowledge of genetic variation within two genes associated with type-
19. Pearson, E. R. Pharmacogenetics and future strategies in treating hyperglycaemia in diabetes. Frontiers in Bioscience 14, 4348-
20. Krueger, G. G. et al. A human interleukin-
9. Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707-
Remaining Questions
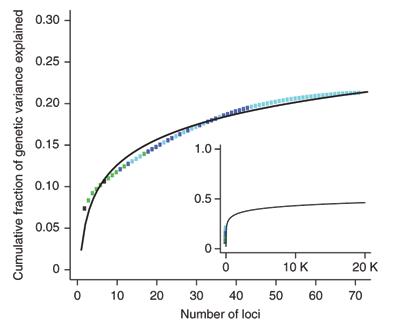
Figure 2 from Genome-
Despite the magnitude and wealth of information provided by both the WTCCC study1 and other recent studies that were even larger in scope, many questions remain about the genetic basis of common diseases. For example, the overall increase in disease risk conferred by the genetic factors identified in the WTCCC study1 and many others is low: approximately 1.2 to 1.5 times (1.33 is the median)13. GWASs also fail to account for all the heritability for any disease or trait. In the case of Crohn's disease, less than 25% of the genetic variation thought to account for its development has been identified to date18 (Figure 4), and in the case of human height, the 180 associated loci that have been identified account for only 10% of its phenotypic variation10. Recent statistical studies suggest, however, that within many GWAS data sets, there are additional signals to uncover that could account for some of the remaining the heritability of traits. For example, scientists estimate that GWASs can uncover at least 33% of the heritability for schizophrenia and that some of this heritability will be shared with bipolar disorder21. In addition, scientists estimate that approximately 20% of heritable variation in height can be discovered with GWASs10.
1. Genome-
13. Hindorff, L. A. et al. Potential etiologic and functional implications of genome-
18. Franke, A. et al. Genome-
10. Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832-
21. Purcell, S. M. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748-
How GWASs Can Translate into Personalized Medicine
How will GWAS findings affect the future of medicine? We are entering era of personalized medicine in which an individual's genetic makeup will eventually determine how his or her therapy is tailored. Therefore, an increased understanding of the genetic basis of common diseases is becoming increasingly critical. It is imperative that we understand how genes predispose individuals to diseases, how disease risk is affected when genes interact with one another, and how rare variants that are difficult to detect using current methods contribute to diseases.
What are some other pressing issues associated with translating GWAS data into personalized medicine? We'd like to know whether different patients can be grouped into subpopulations based on their genetic risk factors, and we'd like to understand the role the environment plays in triggering disease. The Genes, Environment, and Health Initiative of the National Institutes of Health (NIH) (http:/
Summary
Many common human diseases and traits are due in part to common genetic variants. Currently, GWASs are being used to identify these genetic variants. This powerful approach relies on the investigation of large populations of diseased individuals and control individuals from the same population with expansive numbers of DNA markers to identify most of the common genetic variants in the human genome. Over the last few years, more than six hundred GWASs have been performed, leading to the identification of more than seven hundred genetic regions associated with a wide range of human diseases. Despite these successes and the identification of novel pathways for drug development, less than 30% of the genetic contribution to disease has been explained to date. Over the next few years, GWASs are likely to be replaced by complete genomic sequencing, which should help explain the remaining heritability. Progress in identifying genetic risk factors for common diseases and traits is expected to continue at a rapid pace and will undoubtedly lead to exciting new discoveries that will positively affect disease prevention and treatment.
References
1. Genome-
2. Billington, B. P. Gastric cancer; relationships between ABO blood-
3. Carlson, C. S. et al. Selecting a maximally informative set of single-
4. A haplotype map of the human genome. Nature 437, 1299–
5. Todd, J. A. et al. Robust associations of four new chromosome regions from genome-
6. Zeggini, E. et al. Replication of genome-
7. Saxena, R. et al. Genome-
8. D'Addabbo, A. et al. Discovering genetic variants in Crohn's disease by exploring genomic regions enriched of weak association signals. Digestive and Liver Disease [Epub ahead of print] (2011). doi: 10.1016/j.dld.2011.02.010
9. Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707-
10. Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832-
11. Speliotes, E. K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics 42, 937-
12. Manolio, T. A. Genomewide association studies and assessment of the risk of disease. New England Journal of Medicine 363, 166-
13. Hindorff, L. A. et al. Potential etiologic and functional implications of genome-
14. Libioulle, C. et al. Novel Crohn disease locus identified by genome-
15. Klein, R. J. et al. Complement factor H polymorphism in age-
16. Sobrin, L. et al. Genetic profile for five common variants associated with age-
17. Budarf, M. L. et al. GWA studies: Rewriting the story of IBD. Trends in Genetics 25, 137-
18. Franke, A. et al. Genome-
19. Pearson, E. R. Pharmacogenetics and future strategies in treating hyperglycaemia in diabetes. Frontiers in Bioscience 14, 4348-
20. Krueger, G. G. et al. A human interleukin-
21. Purcell, S. M. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748-
Quiz
Question 2.1
1. Why would a GWAS using a larger number of SNPs be more valuable than a study utilizing fewer SNPs?
| A. |
| B. |
| C. |
| D. |
Question 2.2
2. A GWAS identified the complement factor H (CFH) gene as associated with age-related macular degeneration (AMD). Would you expect a person with wild-type CFH to develop AMD? Why or why not?
| A. |
| B. |
| C. |
| D. |
Question 2.3
3. In what way was the GWAS of age-related macular degeneration (AMD) particularly helpful?
| A. |
| B. |
| C. |
| D. |
Question 2.4
4. Personalized medicine strives to treat individual patients best according to the information available about that patient. How can results from a GWAS aid the field of personalized medicine?
| A. |
| B. |
| C. |
Question 2.5
5. Which of the following resources is NOT required to perform a GWAS?
| A. |
| B. |
| C. |
| D. |