Prerequisite skills needed:
- DNA Structure and Base Pairing
- Phosphodiester Bond Formation
- Relevant book section: pages 301-302
By completing this simulation, you will:
- Understand the chemical reactions of PCR
- Carry out PCR by designing primers, mixing components, and analyzing results
- Understand how PCR is used and why it is important to a variety of fields, including the medical and criminal investigative fields
Introduction to BRCA2 and the Polymerase Chain Reaction
There were nearly 1.7 million new breast cancer cases diagnosed worldwide in 2012. In 2015, it is estimated that over 230,000 women in the United States will be diagnosed with invasive breast cancer. 5% to 10% of these cases will occur in women who have inherited a genetic predisposition to breast cancer.
All humans are born with two copies, or alleles, of the BRCA2 gene. If one of these alleles contains a harmful mutation, a person’s lifetime risk of developing breast cancer is greatly increased. About 12% of women in the general population will develop breast cancer during their lifetime. But approximately 45% of women who inherit a harmful BRCA2 mutation will develop breast cancer by age 70. BRCA2 mutations are also linked to increased risk for ovarian and lung cancers.
Courtesy CNN
Introduction to BRCA2 and the Polymerase Chain Reaction
The BRCA2 gene has 27 exons, extends over a region of about 70,000 base pairs on chromosome 13, and encodes a protein with 3418 amino acids. BRCA2 encodes a tumor suppressor protein that functions in repairing damaged DNA. Click the BRCA2 Sequence button above to view the coding sequence of the gene.
You are a molecular biology graduate student. You are studying a tissue sample from a breast cancer tumor, and are trying to determine if the tumor tissue harbors mutations in the BRCA2 gene. Your previous work with the tissue sample leads you to believe that there is a mutation in a region of exon 3 of the BRCA2 gene.
You want to sequence this area of the tumor’s BRCA2 exon 3, so you can identify the mutation. To generate enough sample DNA to use in sequencing, you must amplify the desired region of the tumor genome using the Polymerase Chain Reaction (PCR).
PCR is a laboratory technique that harnesses the power of DNA polymerase enzymes. With just a tiny sample of DNA that contains the region to be amplified, a pair of synthetic oligonucleotides called primers, and a few other reactants in a test tube, a specific fragment of DNA can be multiplied a millionfold in just a few hours.
One particular advantage of PCR is that it is not necessary to know the complete sequence of the DNA segment of interest. As long as some sequence at the end portions of the DNA segment are known, it can be amplified.
PCR—Introduction to Primer Design
The sequence below, which you obtained from a computer database, is a segment of the BRCA2 gene. The portion of BRCA2 exon 3 you wish to amplify from the tumor sample is highlighted in blue.
The first step in PCR is to design two oligonucleotide primers. Primer sequences are carefully selected so that each primer will anneal to a different strand of the double-stranded DNA template, and will allow polymerase to synthesize DNA across the region of interest.
The primers you design should each be 20 nucleotides in length, and should anneal to the sequences immediately adjacent to the blue region of interest.
After a PCR is complete, a portion of the product is usually run on a gel, to check if a DNA fragment of the predicted size was synthesized.
Click Next Page to continue on to the PCR experiment. You will choose the sequence of your primers in a moment.
TGGTTTGAAGAACTTTCTTCAGAAGCTCC
ACCCT
The PCR Reaction
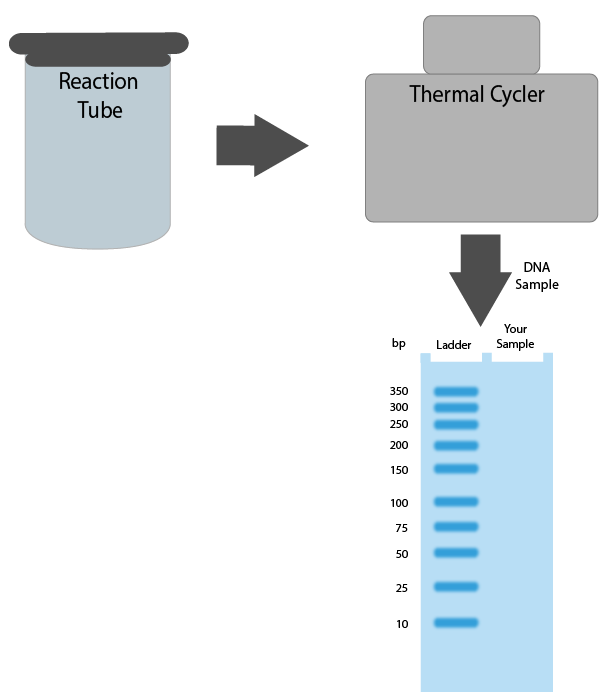







It’s time to set up your PCR experiment:
- Drag-and-drop the components required for PCR into the reaction tube.
- Program the three steps of the thermal cycler by selecting the correct order of events and temperatures from the drop down menus.
- Place the DNA fragment (blue gel band at right) onto the gel, at the position that would indicate your PCR generated a DNA fragment of the desired size. You may need to click back to Section 3 to review the sequence being amplified.
Incorrect - try again.
Incorrect - try again.
Incorrect - try again.
Ordering PCR Primers
CDO Biotechnologies is the company your lab uses to obtain PCR primers. The company synthesizes customized oligonucleotides (oligos) when provided with sequence information from the customer. Enter your primer sequences into their online order form.
Remember: The primers you design should each be 20 nucleotides in length, and should anneal to the sequences immediately adjacent to the blue region of interest.
TGGTTTGAAGAACTTTCTTCAGAAGCTCC
ACCCT
Enter Oligo 1:
Cost: $10 You did not include the appropriate number of nucleotides.
Your primer contains non-nucleotide letters.
Enter Oligo 2:
Cost: $10 You did not include the appropriate number of nucleotides.
Your primer contains non-nucleotide letters.
Cost: $10 You did not include the appropriate number of nucleotides.
Your primer contains non-nucleotide letters.
Enter Oligo 2:
Cost: $10 You did not include the appropriate number of nucleotides.
Your primer contains non-nucleotide letters.
Thank you for purchasing your oligonucleotides from CDO Biotechnologies!
PCR Results
You receive your primers from CDO Biotechnologies, and use them in PCR to amplify the BRCA2 exon 3 fragment from the tumor tissue sample. You run a small sample of the PCR product on a gel, and get the following result:


In analyzing the gel result, which of the following do you agree with?
Click the green button to view a simulation of the PCR you just performed.
You read the gel incorrectly.
Fortunately for your lab, your primer design was correct and your gel shows that your PCR was successful in amplifying the BRCA2 exon 3 fragment. Your advisor is very pleased (although she does wish you would get a little better at reading gels…).
Click the green button to view a simulation of the PCR you just performed.
Unfortunately, your primer design was incorrect and your gel shows that your PCR did not amplify the BRCA2 exon 3 fragment. Your advisor is not thrilled with the fact that you wasted $20 and a bunch of time.
Click the green button to view a simulation of what the PCR would have looked like, had your primer design been correct.
Unfortunately, your primer design was also incorrect and your gel shows that your PCR did not amplify the BRCA2 exon 3 fragment. Your advisor is not thrilled with the fact that you wasted $20 and a bunch of time.
Click the green button to view a simulation of what the PCR would have looked like, had your primer design been correct.
Original DNA Strand:
TGGTTTGAAGAACTTTCTTCAGAAGCTCC
ACCCT
| Correct Primer: | |
| Correct Primer: |
Results
Congratulations! You read the gel correctly and designed the primers correctly, successfully amplifying the BRCA2 exon 3 fragment. Your advisor is very pleased!Click the green button to view a simulation of the PCR you just performed.
Original DNA Strand:
TGGTTTGAAGAACTTTCTTCAGAAGCTCC
ACCCT
| Correct Primer: | |
| Correct Primer: |
Fortunately for your lab, your primer design was correct and your gel shows that your PCR was successful in amplifying the BRCA2 exon 3 fragment. Your advisor is very pleased (although she does wish you would get a little better at reading gels…).
Click the green button to view a simulation of the PCR you just performed.
Original DNA Strand:
TGGTTTGAAGAACTTTCTTCAGAAGCTCC
ACCCT
| Your Primer: | |
| Your Primer: | |
| Correct Primer: | |
| Correct Primer: |
Results
You read the gel correctly. Nice job!Unfortunately, your primer design was incorrect and your gel shows that your PCR did not amplify the BRCA2 exon 3 fragment. Your advisor is not thrilled with the fact that you wasted $20 and a bunch of time.
Hints:
- DNA is complementary.
- The primers need to be adjacent to the target.
- DNA is directional.
- It looks like you may have accidentally entered the same oligo twice. The two primers will most likely have different sequences.
Original DNA Strand:
TGGTTTGAAGAACTTTCTTCAGAAGCTCC
ACCCT
| Your Primer: | |
| Your Primer: | |
| Correct Primer: | |
| Correct Primer: |
Results
You read the gel incorrectly.Unfortunately, your primer design was also incorrect and your gel shows that your PCR did not amplify the BRCA2 exon 3 fragment. Your advisor is not thrilled with the fact that you wasted $20 and a bunch of time.
Hints:
- DNA is complementary.
- The primers need to be adjacent to the target.
- DNA is directional.
- It looks like you may have accidentally entered the same oligo twice. The two primers wil most likely have different sequences.
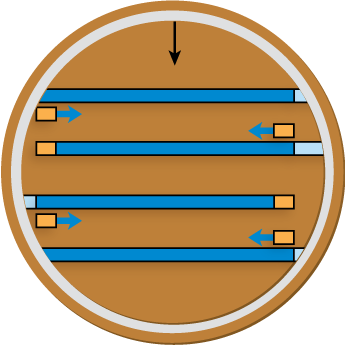
PCR Simulation
Now that you’ve been viewing the PCR simulation, show that you haven’t been asleep. What is the next step in this PCR? Use the selection widget below to choose.
Maybe last time was beginner’s luck. Can you bring it again? Use the selection widget below to choose the next step in this PCR.
It seems that you do know PCR! So just one more time, use the selection widget to specify the next PCR step.
We’ve just completed 4 cycles of PCR. How many DNA fragments will be present after the 5th cycle is complete? Input your answer below, and click Submit.
Correct!
Incorrect, answer is 32 or 25
Click on the Identify Mutation button below to finish the PCR activity.
BRCA2 Mutation Analysis
Exon 3 of BRCA2 Gene:
So to identify the BRCA2 mutation:
- Move the view port left or right until you see a sequence with all red letters except for one (also, the blue bar will turn red).
- Stop moving the view port, and click on the mutated base in the PCR Amplified Fragment box.
Hint: slide slowly and watch carefully!