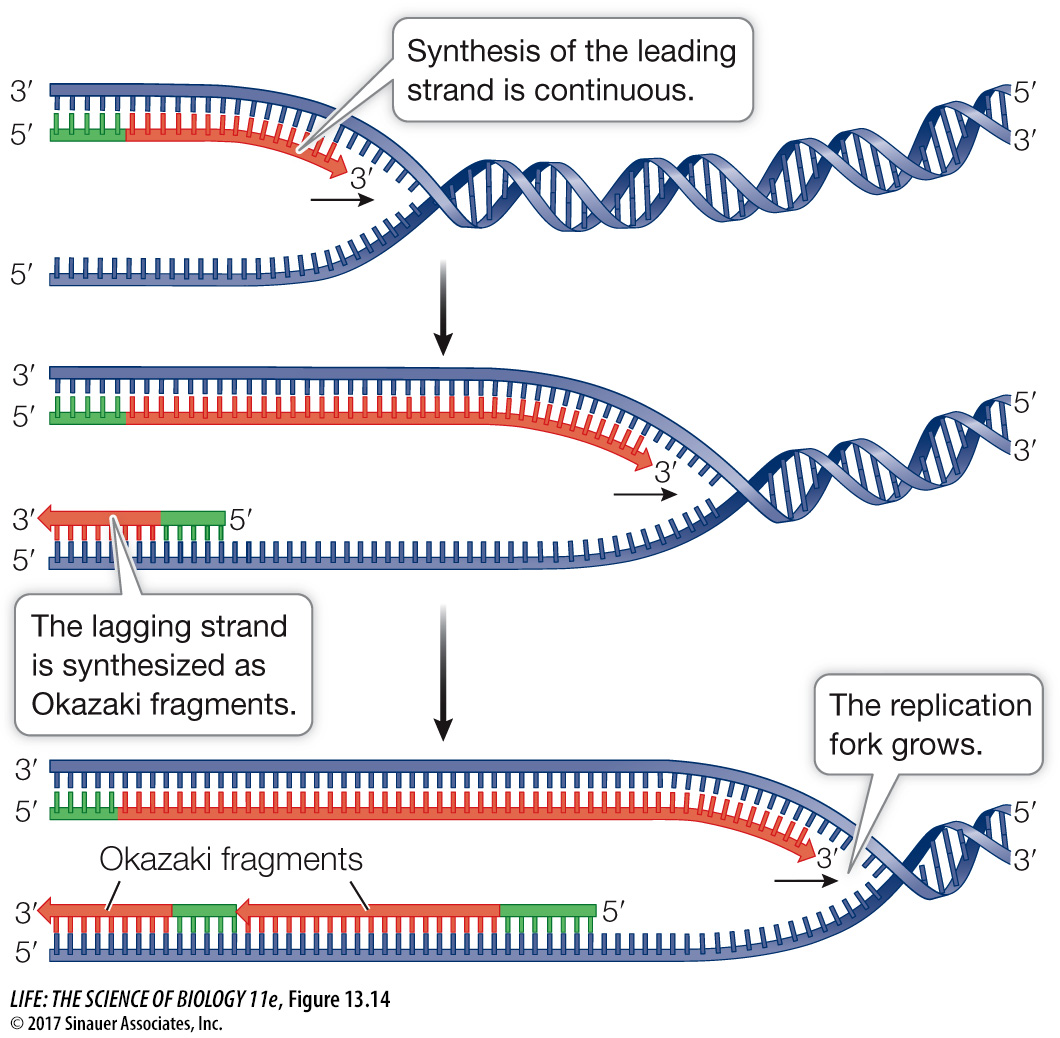
The two DNA strands grow differently at the replication fork
The DNA at the replication fork—
Animation 13.4 Leading and Lagging Strand Synthesis
One newly replicating strand (the leading strand) is oriented so that it can grow continuously at its 3′ end as the fork opens up.
The other new strand (the lagging strand) is oriented so that as the fork opens up, its exposed 3′ end gets farther and farther away from the fork, and an unreplicated gap is formed. This gap would get bigger and bigger if there were not a special mechanism to overcome this problem.
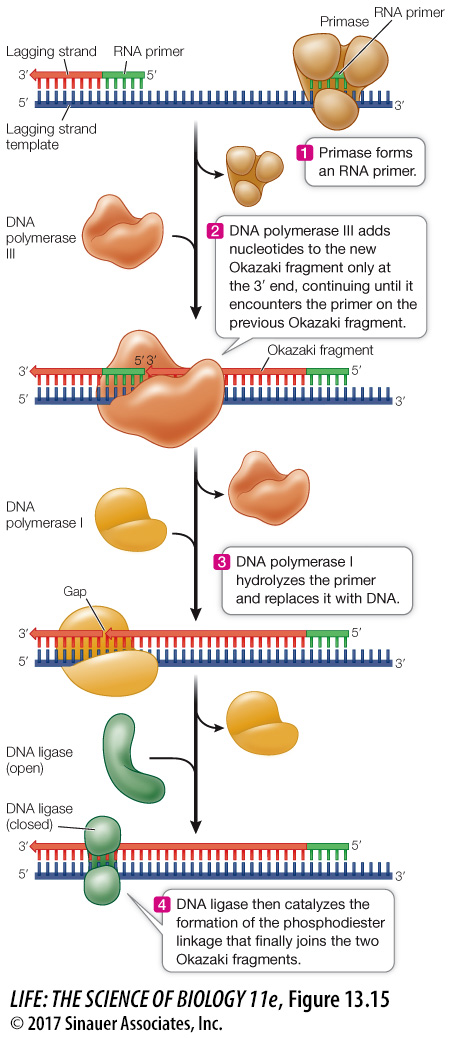
Synthesis of the lagging strand requires the synthesis of relatively small, discontinuous stretches of DNA (100–
A single primer is needed for synthesis of the leading strand, but each Okazaki fragment requires its own primer to be synthesized by the primase. In bacteria, DNA polymerase III then synthesizes an Okazaki fragment by adding nucleotides to one primer until it reaches the primer of the previous fragment (Figure 13.15). At this point, DNA polymerase I removes the old primer and replaces it with DNA. Left behind is a tiny nick—
DNA replication involves remarkable teamwork among various proteins that act on the DNA strands. Let’s review the proteins involved in DNA replication in the order of their activity at the replication fork:
DNA helicase unwinds the double helix and separates the two strands.
Single-
strand binding proteins bind to separated strands and prevent them from re-forming the double helix. DNA primase makes RNA primers.
DNA polymerase links new nucleotides to form the new DNA strands and removes the primers.
DNA ligase connects Okazaki fragments made by DNA polymerase to one another.
Working together, these proteins make new DNA at a rate in excess of 1,000 base pairs per second, committing errors in fewer than 1 base in a million.
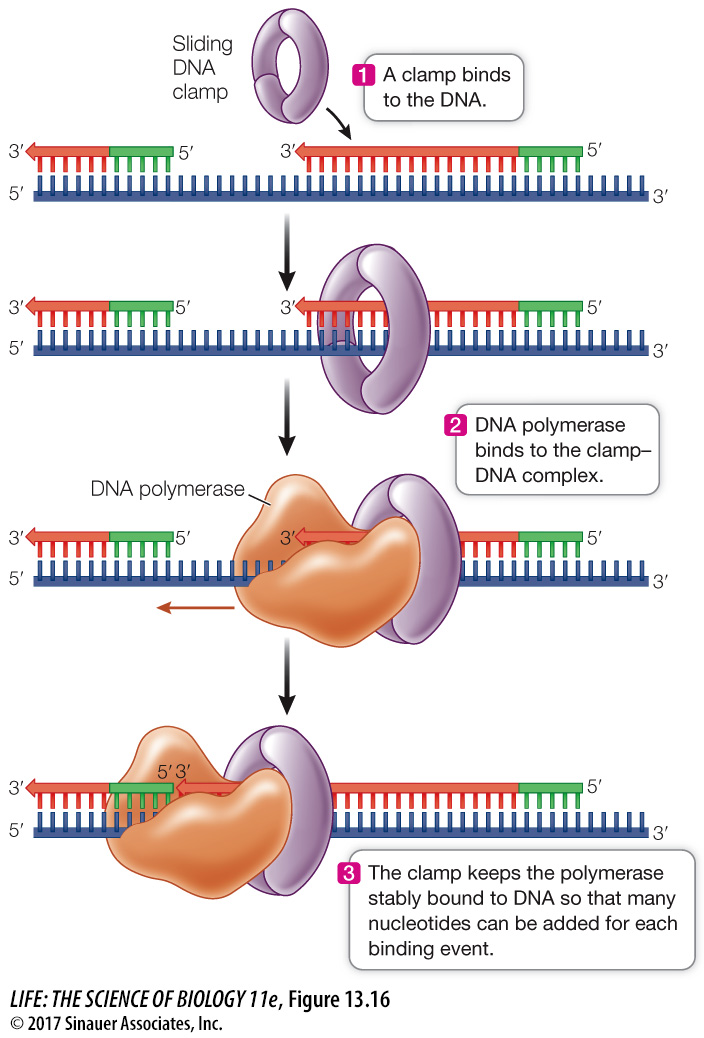
A SLIDING CLAMP INCREASES THE RATE OF DNA REPLICATION How do DNA polymerases work so fast? We saw in Key Concept 8.3 that an enzyme catalyzes a chemical reaction:
Substrate binds to enzyme → one product is formed → enzyme is released → cycle repeats
DNA replication would not proceed as rapidly as it does if it went through such a cycle for each nucleotide. Instead, DNA polymerases are processive—that is, they catalyze the formation of many phosphodiester linkages each time they bind to a DNA molecule:
Substrates bind to enzyme → many products are formed → enzyme is released → cycle repeats
The DNA polymerase–
DNA IS THREADED THROUGH A REPLICATION COMPLEX So far, you are probably envisioning DNA replication as a locomotive (the replication complex) moving along a railroad track (the DNA). But this is not so. Commonly in eukaryotes, the replication complexes seem to be stationary, attached at specific positions in the nucleus. It is the DNA that moves, essentially sliding into the replication complex as one double-