Expanding triplet repeats demonstrate the fragility of some human genes
About one-fifth of all males who have the fragile-X chromosomal abnormality are phenotypically normal, as are most of their daughters. But many of those daughters’ sons are intellectually disabled. In a family in which fragile-X syndrome appears, later generations tend to show earlier onset and more severe symptoms of the disease. It is almost as if the abnormal allele itself is changing—and getting worse. And that’s exactly what is happening.
The gene associated with fragile-X syndrome (FMR1) contains a repeated triplet, CGG, at a certain point in the promoter region (Figure 15.9). In normal people, this triplet is repeated 6 to 54 times (the average is 29). In intellectually disabled people with fragile-X syndrome, the CGG sequence is repeated 200 to 2,000 times.
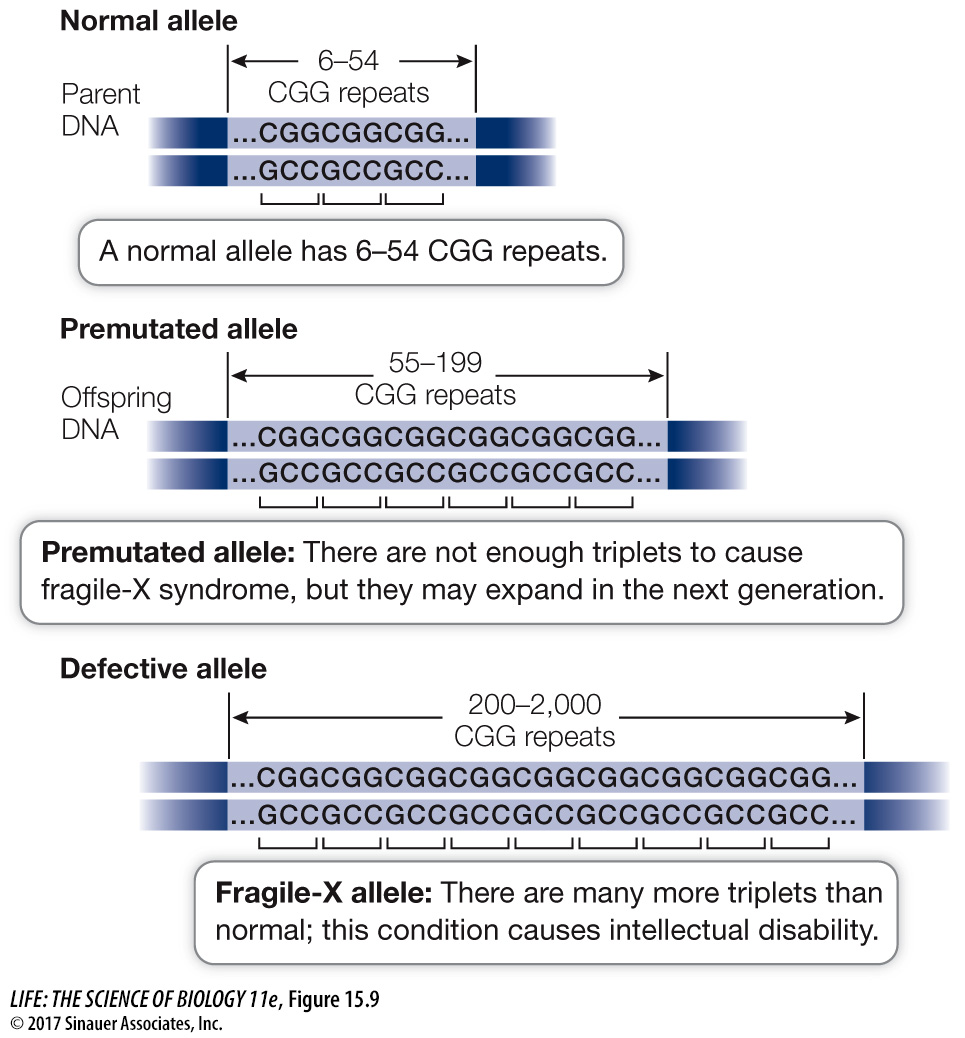
Figure 15.9 The CGG Repeats in the FMR1 Gene Expand with Each Generation The genetic defect in fragile-X syndrome is caused by 200 or more repeats of the CGG triplet.
Males carrying a moderate number of repeats (55–199) show no symptoms and are called premutated. These repeats become more numerous as the daughters of these men pass the chromosome on to their children. With 200 or more repeats, increased methylation of the cytosines in the CGG triplets is likely, which inhibits transcription of the FMR1 gene. The normal role of the protein product of this gene is to bind to mRNAs involved in neuron function and to regulate their translation at the ribosome. When the FMR1 protein is not made in adequate amounts, these mRNAs are not properly translated, and nerve cells die. Their loss often results in intellectual disability. The methylated cytosines bind proteins that cause the chromosome constriction and fragile appearance.
This phenomenon of expanding triplet repeats has been found in more than a dozen other diseases, such as myotonic dystrophy (involving repeated CTG triplets) and Huntington’s disease (in which CAG is repeated). Such repeats, which may be found within a protein-coding region or outside it, appear to be present in many other genes without causing harm. How the repeats expand is not known; one hypothesis is that DNA polymerase may slip after copying a repeat and then fall back to copy it again.
