Histone protein modifications affect transcription
Another mechanism for epigenetic gene regulation is the alteration of chromatin structure, or chromatin remodeling. As we saw in Chapter 11, DNA is packaged with histone proteins into nucleosomes (see Figure 11.8), which can make DNA physically inaccessible to RNA polymerase and the rest of the transcription apparatus. Each histone protein has a “tail” of approximately 20 amino acids at its N terminus that sticks out of the compact structure and contains certain positively charged amino acids (notably lysine). Ordinarily there is strong ionic attraction between the positively charged histone proteins and DNA, which is negatively charged because of its phosphate groups. However, enzymes called histone acetyltransferases can add acetyl groups to these positively charged amino acids, thus changing their charges:
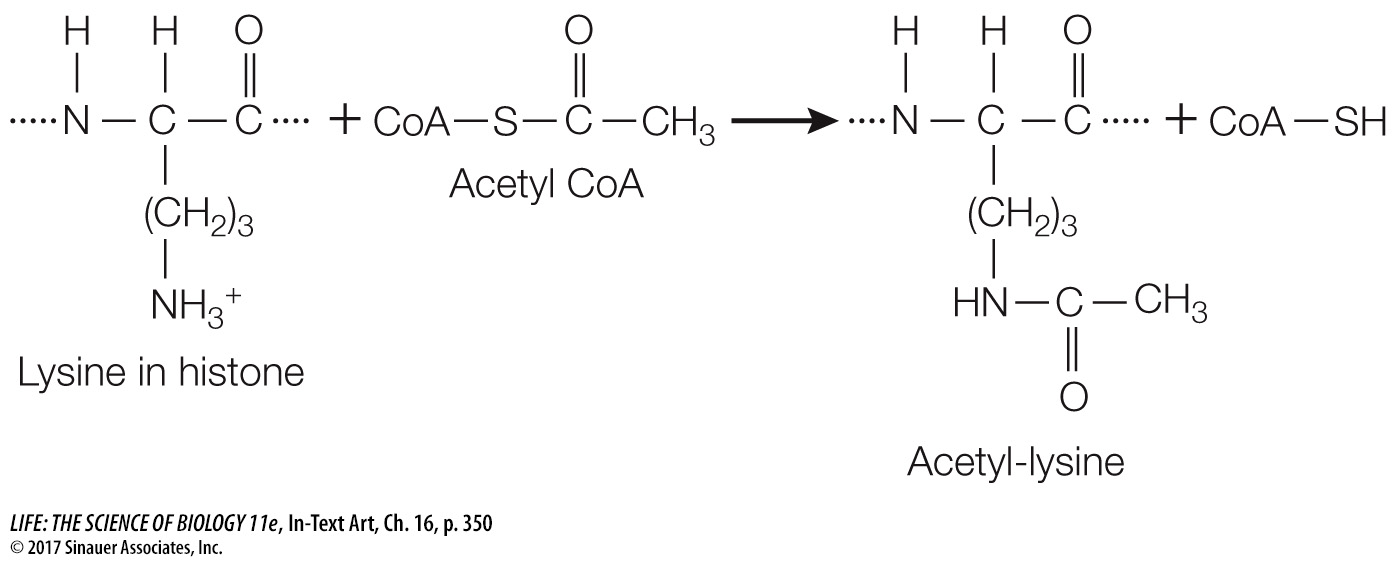
Reducing the positive charges of the histone tails reduces the affinity of the histones for DNA, opening up the compact nucleosome (Figure 16.15). Additional chromatin remodeling proteins can bind to the loosened nucleosome–
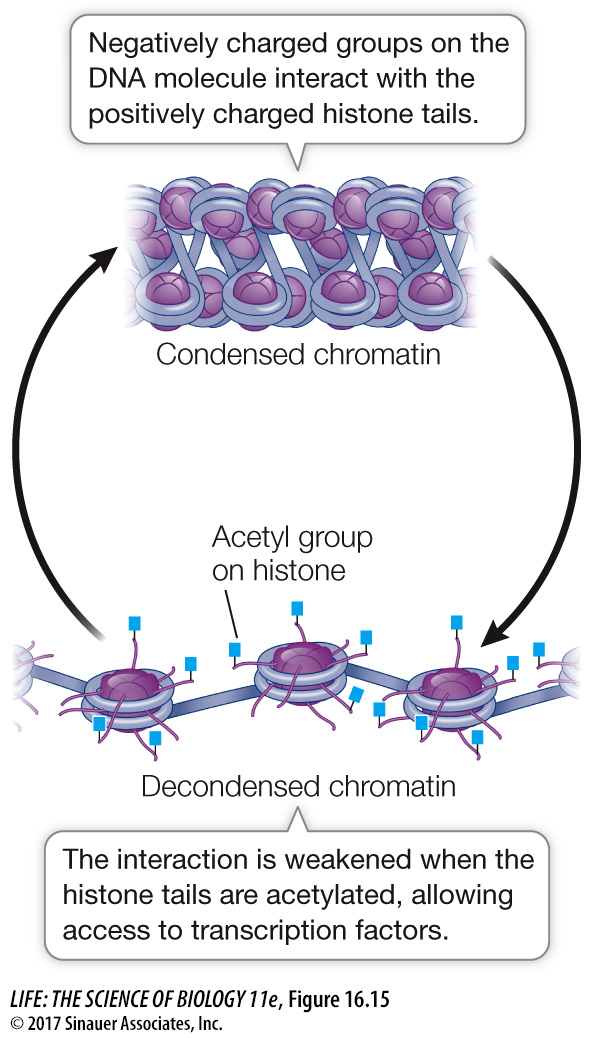
Another kind of chromatin remodeling protein, histone deacetylase, can remove the acetyl groups from histones and thereby repress transcription. Histone deacetylases are targets for drug development to treat some forms of cancer. As noted above, certain genes block cell division in normal specialized tissues. In some cancers these genes are less active than in normal cells, and the histones near them show excessive levels of deacetylation. Theoretically, a drug acting as a histone deacetylase inhibitor could tip the balance toward acetylation, and this might activate genes that normally inhibit cell division.
Acetylation isn’t the only type of histone modification that can affect gene activation and repression. For example, histone methylation (not to be confused with DNA methylation) is associated with gene inactivation, and histone phosphorylation also affects gene expression, the specific effects depending on which amino acids are modified. All of these effects are reversible, and so the activity of a eukaryotic gene may be determined by very complex patterns of histone modification.