recap
16.4 recap
Epigenetics describes stable changes in gene expression that do not involve changes in DNA sequences. These changes involve modifications of DNA (cytosine methylation) or of histone proteins bound to DNA. Epigenetic changes can be affected by the environment. Large stretches of DNA can be epigenetically modified, leading to inactivtion of many genes.
learning outcomes
You should be able to:
Describe the role of methylation in gene expression.
Explain how acetylation of proteins changes the structure of chromatin and affects the rate of transcription.
Cite evidence supporting the claim that environmental effects have a strong role in epigenetic changes.
Describe how and where X chromosome inactivation occurs, and explain why it is believed to occur.
1.
How do histone modifications affect transcription?
Histone proteins are positively charged and bind to DNA, generally blocking transcription. Acetylation of histones neutralizes the positive charge and thus the histones do not bind to DNA as tightly, which opens up the chromatin structure for transcription. By contrast, histone deacetylation removes acetyl groups, restoring positive charges on histones so transcription is repressed.
2.
What is the evidence that epigenetic modifications affect behavior?
See the opening story and Investigating Life: Gene Expression and Behavior (p. 351). The behavioral environment appears to alter DNA epigenetically by increasing DNA methylation at promoters in genes in brain tissues, thus changing the rate of gene transcription.
3.
How does X chromosome inactivation occur, and why is it believed to occur?
X chromosome inactivation is shown in Figure 16.16. The Xist gene on the X chromosome is transcribed to make a short RNA that binds to the rest of the X chromosome, inhibiting transcription of the other genes. Chromosome proteins bind to the inactive X chromosome, causing heterochromatin to form and inhibiting gene expression. X chromosome inactivation is believed to occur in order to balance the expression of X-
4.
In colorectal cancer, certain tumor suppressor genes are not active, and this results in uncontrolled cell division. Two possible explanations for the inactive genes are (a) mutations in the coding regions, resulting in inactive proteins, and (b) epigenetic silencing at the promoters of the genes, resulting in reduced transcription. How would you investigate these two possibilities?
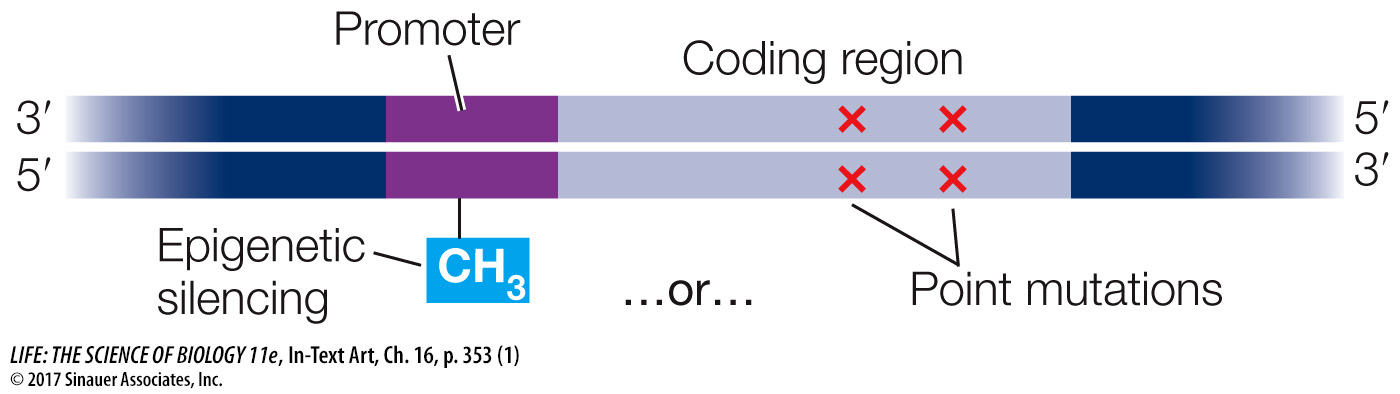
You could sequence the relevant genes of colorectal cancer cells and look for mutations that lead to aberrant function, then isolate the proteins involved and determine that their functions are indeed abnormal. To show epigenetic silencing, you might sequence the promoters of the genes and look for epigenetic changes (e.g., cytosine methylation, which would be increased if there were transcriptional silencing). Then you could examine the tumor cells to see if the active proteins were there but in small amounts.
Gene expression involves transcription and then translation. So far we have described how gene expression is regulated at the transcriptional level. But as Figure 16.7 shows, there are many points at which regulation can occur after the initial gene transcript is made.