Eukaryotes have gene families
For about half of all eukaryotic protein-
*connect the concepts Several specific examples of gene duplication resulting in gene homologs are discussed in Chapter 19, such as the evolution of remarkable homology across animal species of the Hox genes, which regulate early development.
The DNA sequences in a gene family are usually different from one another. As long as at least one member encodes a functional protein, the other members may mutate in ways that change the functions of the proteins they encode. During evolution, the availability of multiple copies of a gene allows for selection of mutations that provide advantages under certain circumstances. If a mutated gene is useful, it may be selected for in succeeding generations. If the mutated gene is a total loss, the functional copy is still there to carry out its role.
Let’s take a look at one gene family, the globin genes in vertebrates. The globins are components of hemoglobin and myoglobin (an oxygen-
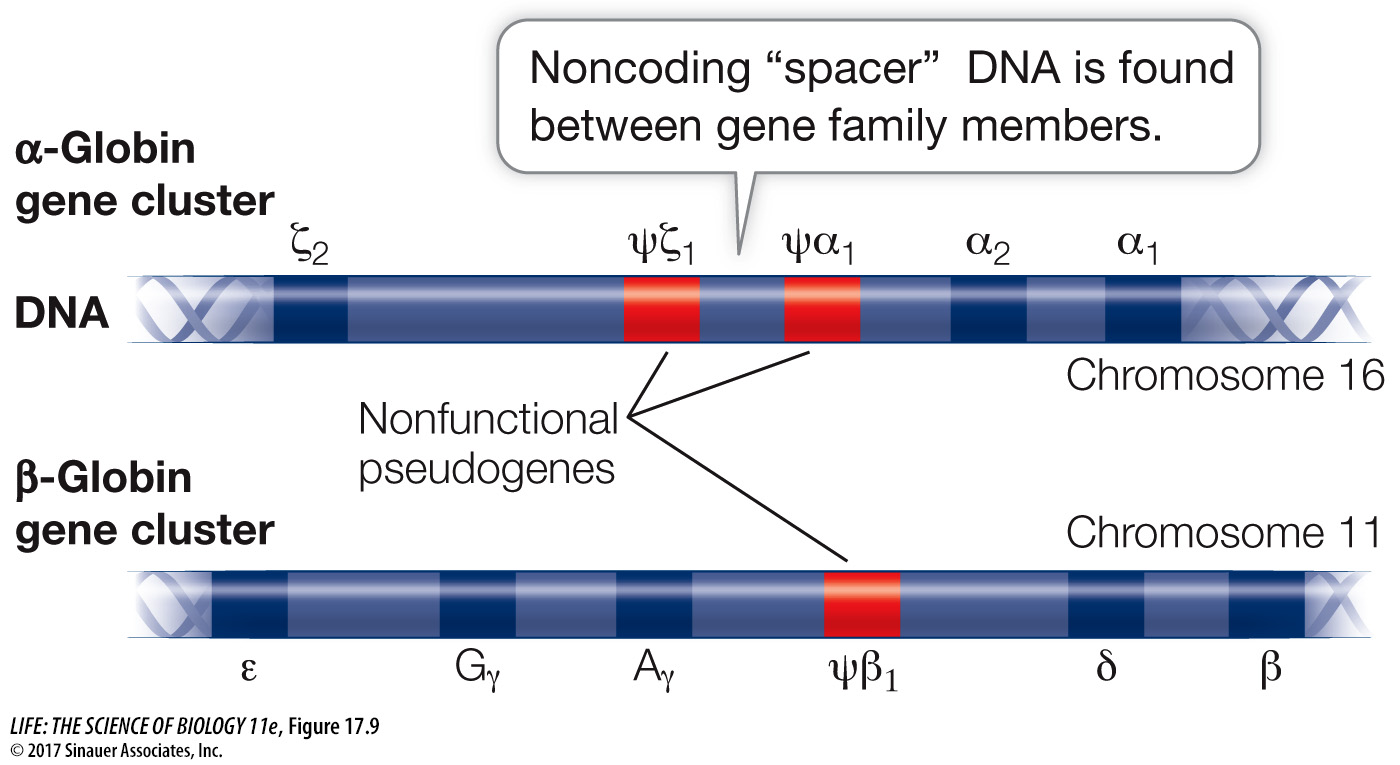
During human development, different members of the globin gene cluster are expressed at different times and in different tissues. This differential gene expression has great physiological significance. For example, hemoglobin that contains γ-globin, a subunit found in the hemoglobin of the human fetus, binds O2 more tightly than adult hemoglobin does. This specialized form of hemoglobin ensures that in the placenta, O2 will be transferred from the mother’s blood to the developing fetus’s blood. Just before birth the liver stops synthesizing fetal hemoglobin and the bone marrow cells take over, making the adult form (2 α and 2 β). Thus hemoglobins with different binding affinities for O2 are provided at different stages of human development.
In addition to genes that encode proteins, many gene families include nonfunctional pseudogenes, which are designated with the Greek letter psi (ψ) (see Figure 17.9). Pseudogenes result from mutations that cause a loss of function rather than an enhanced or new function. The DNA sequence of a pseudogene may not differ greatly from that of other family members. It may simply lack a promoter, for example, and thus fail to be transcribed. Or it may lack a recognition site needed for the removal of an intron, so that the transcript it makes is not correctly processed into a useful mature mRNA. In some gene families pseudogenes outnumber functional genes. In such cases, there appears to be no evolutionary advantage for the deletion of the pseudogenes, even though they have no apparent function.