Genome sequences yield several kinds of information
New genome sequences are being published at an accelerating pace, creating a torrent of biological information. This information is used in two related fields of research, both focused on studying genomes. In functional genomics, biologists use sequence information to identify the functions of various parts of genomes (the meaningful sequences in genomes such as those encoding mRNA, tRNA, and regulatory sequences; see Key Concept 14.4). These parts include:
Open reading frames, which are the coding regions of genes. For protein-
coding genes, these regions can be recognized by the start and stop codons for translation, and by recognition sequences that indicate the locations of introns. A major goal of functional genomics is to understand the function of every open reading frame in each genome. Amino acid sequences of proteins, which can be deduced by applying the genetic code to the DNA sequences of open reading frames.
Regulatory sequences, such as promoters, enhancers, and terminators for transcription. These are identified by their proximity to open reading frames and because they contain recognition sequences for the binding of specific transcription factors.
RNA genes, including rRNA, tRNA, small nuclear RNA, and microRNA genes.
Other noncoding sequences that can be classified into various categories, including centromeric and telomeric regions, transposons, and other repetitive sequences.
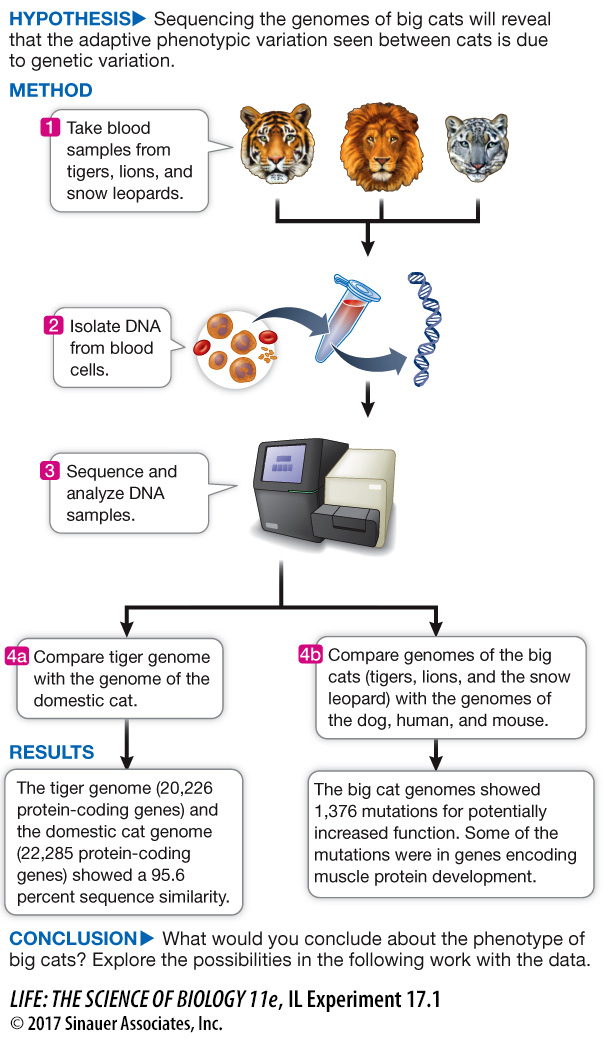
Sequence information is also used in comparative genomics: the comparison of a newly sequenced genome (or parts thereof) with sequences from other organisms. The Dog Genome Project described at the opening of this chapter, for example, has yielded information not only about dogs, but on how the dog genome compares with other animal genomes. Genome comparisons can provide further information about the functions of sequences and can be used to trace evolutionary relationships among different species. Each animal genome sequenced offers new insights. Investigating Life: Comparative Analysis of the Tiger Genome describes the recent sequencing of the tiger genome and how it relates to the genomes of other cats and other mammals.
investigating life
Comparative Analysis of the Tiger Genome
experiment
Original Paper: Cho, Y. S. et al. 2013. The tiger genome and comparative analysis with lion and snow leopard genomes. Nature Communications 4: 1–
Panthera tigris, the tiger, is possibly the most famous endangered species. Fewer than 4,000 individuals remain in the wild. While a century ago there were nine recognized genetically distinct subspecies, four have become extinct. The five extant subspecies include the Bengal tiger seen at zoos and the white Amur tiger that lives in snowy regions of Russia, China, and North Korea. While the genomes of other cats, such as the lion, snow leopard, and domestic cat, had already been sequenced, that of the tiger had not until this study.
work with the data
An international team led by Jong Bhak at the Genome Research Foundation in Suwon, South Korea, determined the sequence of tiger DNA and compared it with lion and snow leopard DNA, as well as that of the domestic cat.
QUESTIONS
1.
The overall sequence of tiger DNA was compared with that of the domestic cat. For reference, a comparison between the human and gorilla genomes was included. Evolutionary distances (last common ancestor) were also estimated. The results are shown in the table. What can you conclude about the rate of evolutionary change between the two cat genomes and between the human and gorilla genomes?
| Comparison groups | Last common ancestor (millions of years ago) |
Genome sequence similarity (%) |
|---|---|---|
| Domestic cat and tiger | 10.8 | 95.6 |
| Human and gorilla | 8.8 | 94.8 |
There was a 4.4 percent change in DNA sequence between the two cat genomes in 10.8 million years. This indicates a rate of change of 0.4 percent per million years. By comparison, the rate of change between the human and gorilla genomes was 0.6 percent per million years, or 50 percent faster.
2.
While there are 20,226 protein-
Looking at the Venn diagram, how many gene families are shared by all the mammalian genomes examined?
How many gene families are unique to the tiger and domestic cat genomes? How many are unique to the human and mouse genomes?
What do these data indicate about the basic mammalian genome?
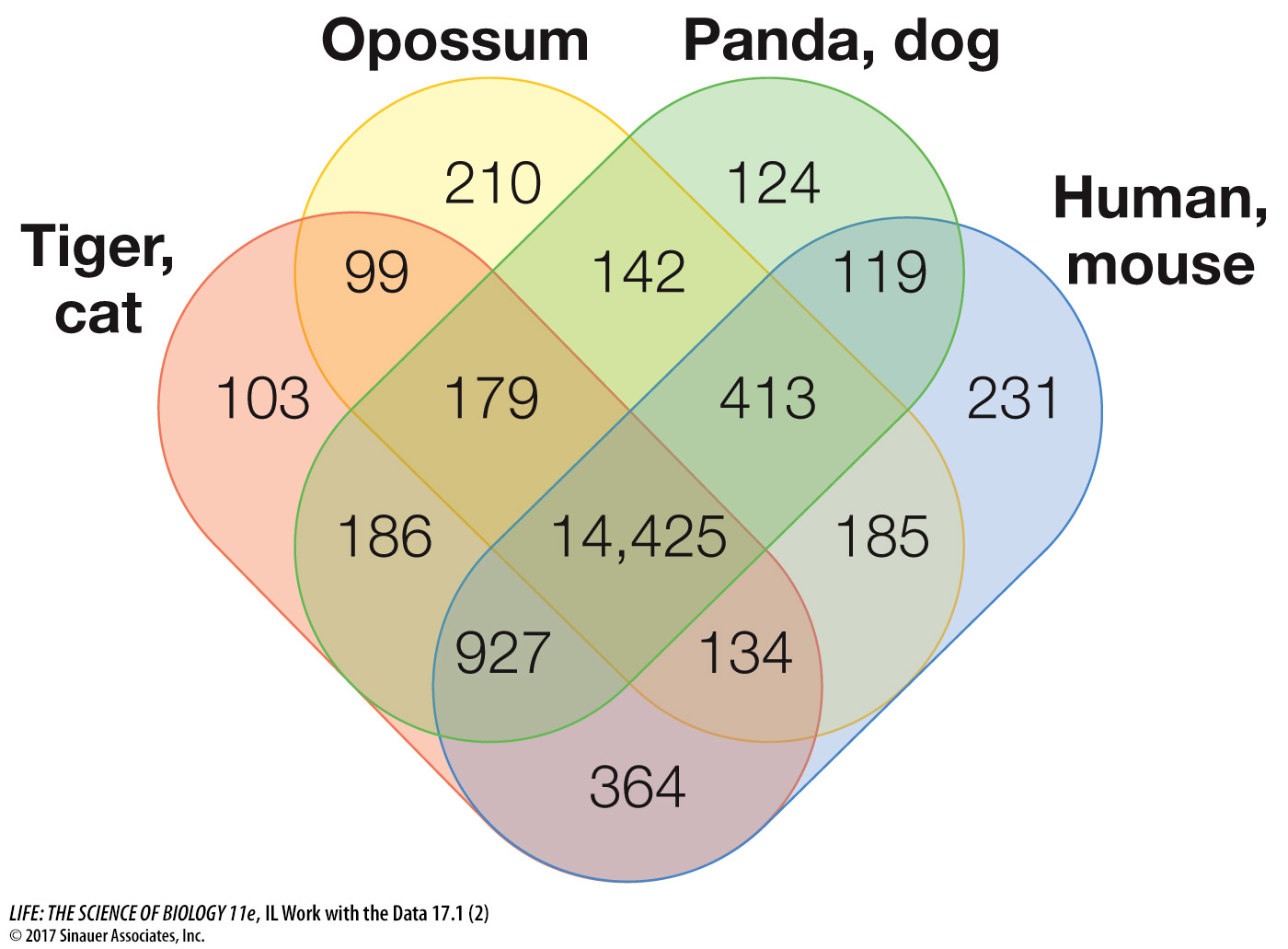
14,425 gene families are shared by all the mammalian genomes examined.
103 are unique to the tiger and domestic cat genomes; 231 are unique to the human and mouse genomes.
Over 90 percent of the mammalian genome is common to all mammals. Relatively few gene families are unique to each kind of mammal examined.
3.
The tiger genome was analyzed for sequences of protein-
Olfactory (smell) receptors: 289 genes
G protein–
Signal transduction: 295 genes
Protein metabolism: 220 genes
What do these genotype data indicate about the phenotype of the tiger?
The tiger is a hunter, and its genome reflects this, with genes for smell, signaling and digestion.
A similar work with the data exercise may be assigned in LaunchPad.