Phylogenies are reconstructed from many sources of data
Naturalists have constructed various forms of phylogenetic trees for more than 150 years. In fact, the only figure in the first edition of On the Origin of Species was a phylogenetic tree. Tree construction has been revolutionized, however, by the advent of computer software that allows us to consider far more data and analyze far more traits than could ever before be processed. Combining these advances in methodology with the massive comparative data sets being generated through studies of genomes, biologists are learning details about the tree of life at a remarkable pace (see Appendix A: The Tree of Life).
Any trait that is genetically determined, and therefore heritable, can be used in a phylogenetic analysis. Evolutionary relationships can be revealed through studies of morphology, development, the fossil record, behavioral traits, and molecular traits such as DNA and protein sequences. Let’s take a closer look at the types of data used in modern phylogenetic analyses.
MORPHOLOGY An important source of phylogenetic information is morphology: the presence, size, shape, and other attributes of body parts. Since living organisms have been observed, depicted, collected, and studied for millennia, we have a wealth of recorded morphological data as well as extensive museum and herbarium collections of organisms whose traits can be measured. New technological tools, such as the electron microscope and computed tomography (CT) scans, enable systematists to examine and analyze the structures of organisms at much finer scales than was formerly possible.
Most species are described and known primarily by their morphology, and morphology still provides the most comprehensive data set available for many taxa. The morphological features that are important for phylogenetic analysis are often specific to a particular group. For example, the presence, development, shape, and size of various features of the skeletal system are important in vertebrate phylogeny, whereas floral structures are important for studying the relationships among flowering plants.
Morphological approaches to phylogenetic analysis have some limitations, however. Some taxa exhibit little morphological diversity, despite great species diversity. For example, the phylogeny of the leopard frogs of North and Central America would be difficult to infer from morphological differences alone, because the many species look very similar, despite important differences in their behavior and physiology. At the other extreme, few morphological traits can be compared across distantly related species (earthworms and mammals, for example). Furthermore, some morphological variation has an environmental (rather than a genetic) basis and so must be excluded from phylogenetic analyses. An accurate phylogenetic analysis often requires information beyond that supplied by morphology.
DEVELOPMENT Similarities in *developmental patterns may reveal evolutionary relationships. Some organisms exhibit similarities only in early developmental stages. The larvae of marine creatures called sea squirts, for example, have a flexible gelatinous rod in the back—
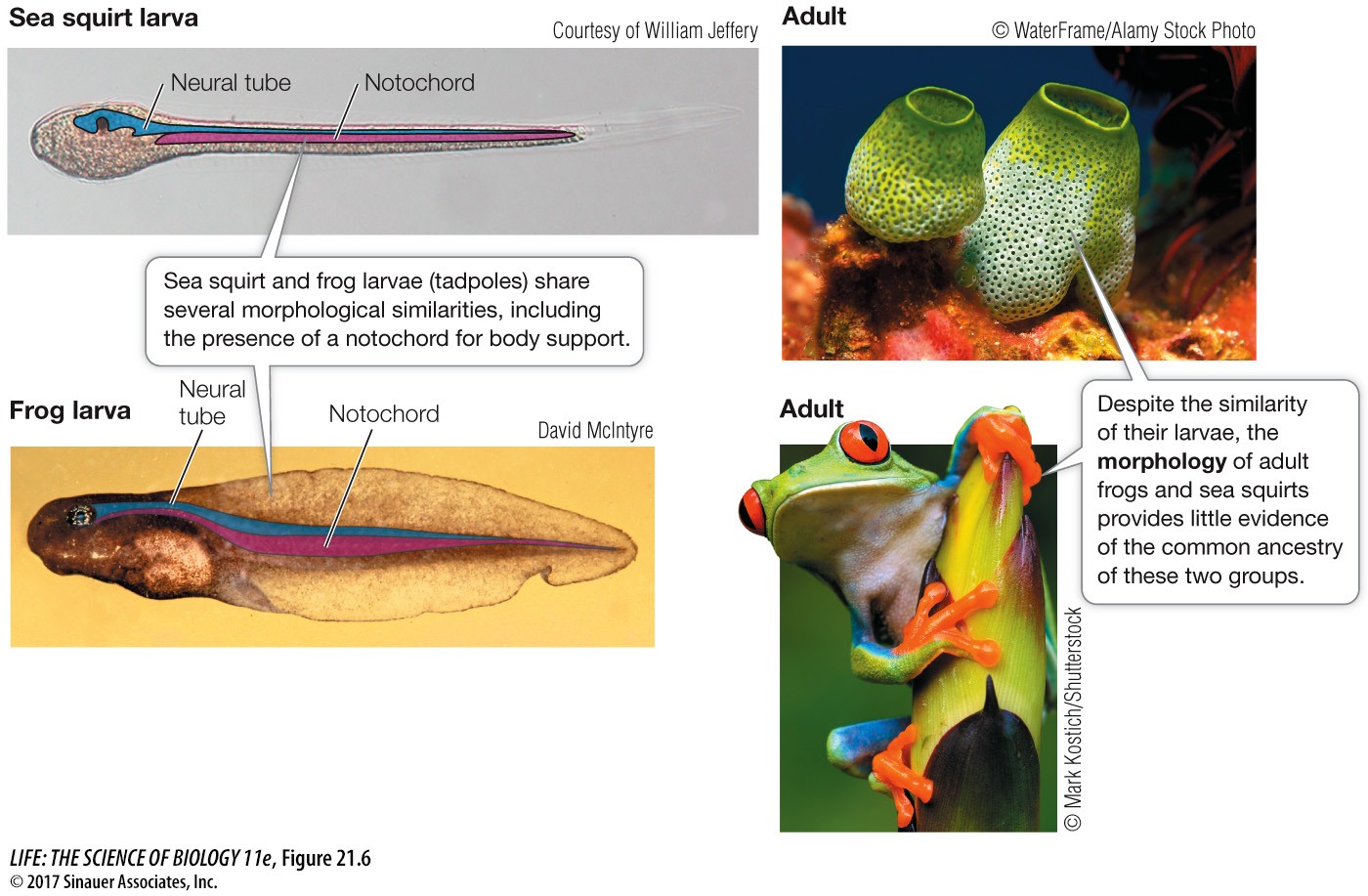
*connect the concepts Evolutionary developmental biology, the subject of Chapter 19, compares the developmental processes of different organisms to determine the ancestral relationship between them, and to discover how developmental processes evolved. Genetic toolkit genes are expressed in different ways in different species, resulting in major morphological differences among species (see Key Concepts 19.4 and 19.5). The existence of highly conserved development genes makes it likely that similar traits will evolve repeatedly.
PALEONTOLOGY The fossil record is another important source of information on evolutionary history. Fossils show us where and when organisms lived in the past and give us an idea of what they looked like. Fossils provide important evidence that helps us distinguish ancestral from derived traits. The fossil record can also reveal when lineages diverged and began their independent evolutionary histories. Furthermore, in groups with few species that have survived to the present, information on extinct species is often critical to an understanding of the large divergences among the surviving species. The fossil record has limitations, however. Few or no fossils have been found for some groups, and the fossil record for many groups is fragmentary.
BEHAVIOR Some behavioral traits are culturally transmitted and others are genetically inherited. If a particular behavior is culturally transmitted, it may not accurately reflect evolutionary relationships (but may nonetheless reflect cultural connections). Many bird songs, for instance, are learned and may be inappropriate traits for phylogenetic analysis. Frog calls, however, are genetically determined and appear to be acceptable sources of information for reconstructing phylogenies.
MOLECULAR DATA All heritable variation is encoded in DNA, and so the complete genome of an organism contains an enormous set of traits (the individual nucleotide bases of DNA) that can be used in phylogenetic analyses. In recent years, DNA sequences have become among the most widely used sources of data for constructing phylogenetic trees. Comparisons of nucleotide sequences are not limited to the DNA in the cell nucleus. Eukaryotes have genes in their mitochondria as well as in their nuclei. Plant cells also have genes in their chloroplasts. The chloroplast genome (cpDNA), which is used extensively in phylogenetic studies of plants, has changed slowly over evolutionary time, so it is often used to study relatively ancient phylogenetic relationships. Most animal mitochondrial DNA (mtDNA) has changed more rapidly, so mitochondrial genes are used to study evolutionary relationships among closely related animal species (the mitochondrial genes of plants evolve more slowly). Many nuclear gene sequences are also commonly analyzed, and now that entire genomes have been sequenced from many species, they too are used to construct phylogenetic trees. Information on gene products (such as the amino acid sequences of proteins) is also widely used for phylogenetic analyses.