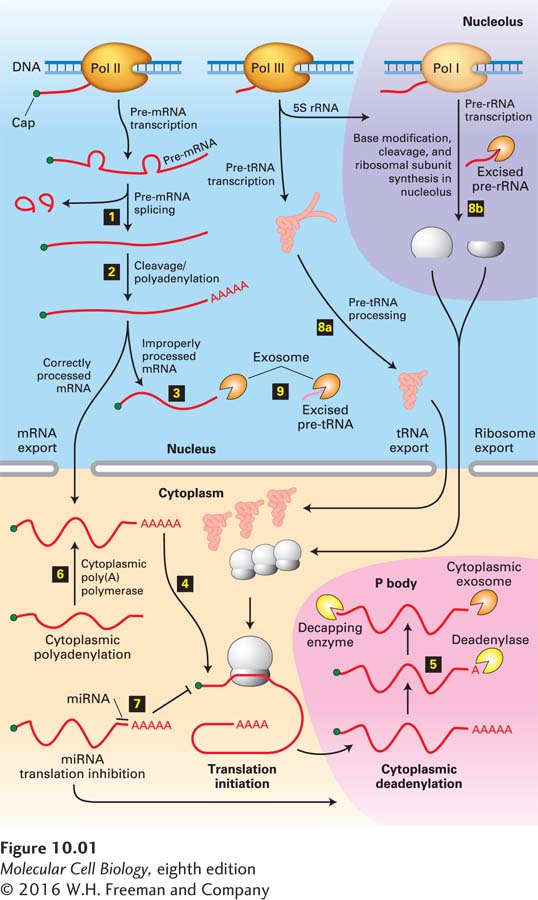
FIGURE 10- t- n- e- 4- 2-