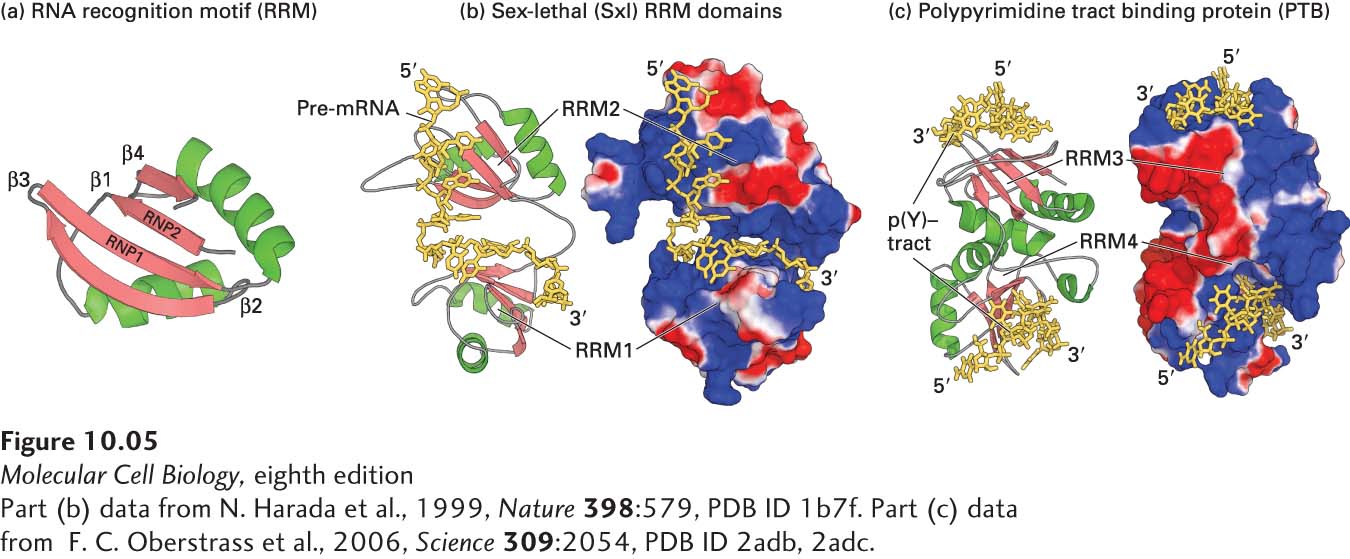
FIGURE 10- 5 Structure of the RRM domain and its interaction with RNA. (a) Ribbon diagram of the RRM domain found in hnRNP proteins, showing the two α helices (green) and four β strands (red) that characterize this motif. The conserved RNP1 and RNP2 regions are located in the two central β strands. (b, c) Ribbon diagram and surface representation of the two RRM domains in Drosophila Sex- lethal (Sxl) protein (b) and the polypyrimidine tract- binding protein (PTB) (c). In both (b) and (c), positively charged regions are shown in shades of blue; negatively charged regions, in shades of red; RNA is yellow. The two RRMs in Sxl are oriented like the two parts of an open pair of castanets, with the β sheets of the RRMs facing toward each other. The pre- mRNA is bound to the surfaces of the positively charged β sheets, making most of its contacts with the RNP1 and RNP2 regions of each RRM. PTB has a strikingly different orientation of RRM domains, illustrating that RRMs are oriented in different relative positions in different hnRNPs. The p(Y)-tract is a polypyrimidine tract. In PTB, the two RRMs associate through their α helices so that the positively charged β sheets face away from each other, upward for RRM3 and downward for RRM4. The structure of CUCUCU single- stranded RNA bound to each of the two RRMs was determined, explaining how PTB can bind to two tracts of six pyrimidines in a single RNA if they are separated by a loop of 15 or more nucleotides. This ability of PTB to form a small loop in a pre- mRNA probably contributes to its ability to function as a splicing repressor at exons where the upstream 3′ splice site or the downstream 5′ splice site is flanked by two polypyrimidine tracts. See K. Nagai et al., 1995, Trends Biochem. Sci. 20:235.
[Part (b) data from N. Harada et al., 1999, Nature 398:579, PDB ID 1b7f. Part (c) data from F. C. Oberstrass et al., 2006, Science 309:2054, PDB ID 2adb, 2adc.]
[Leave] [Close]