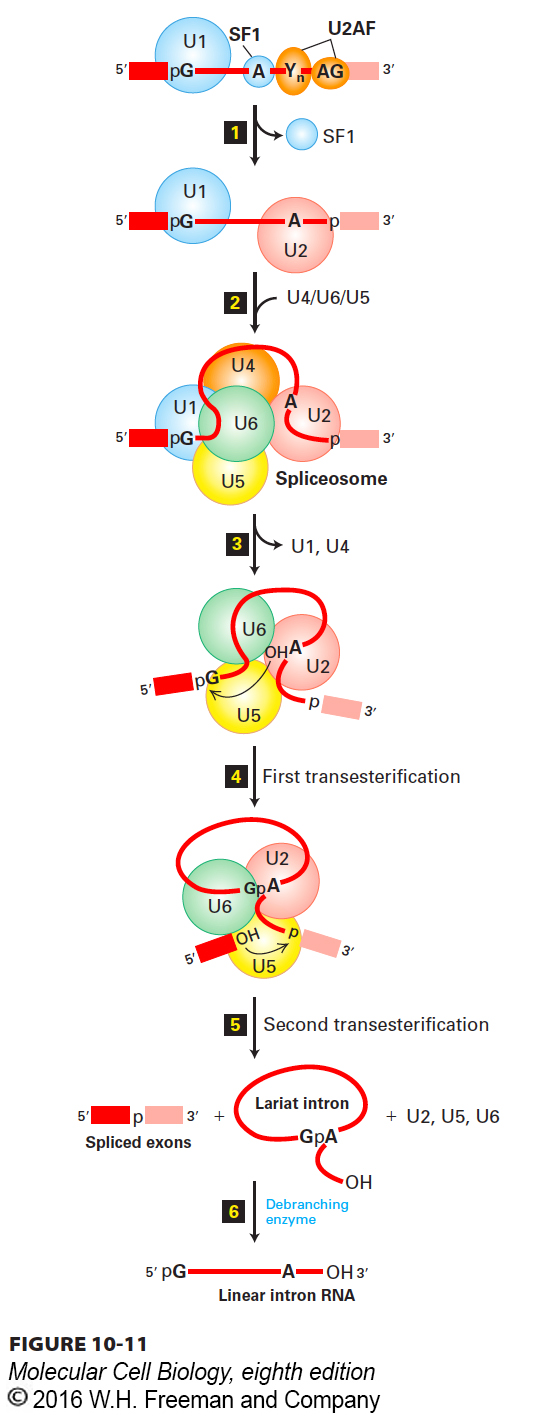
FIGURE 10- e- e- e- h- h- e- 0- e- 0-