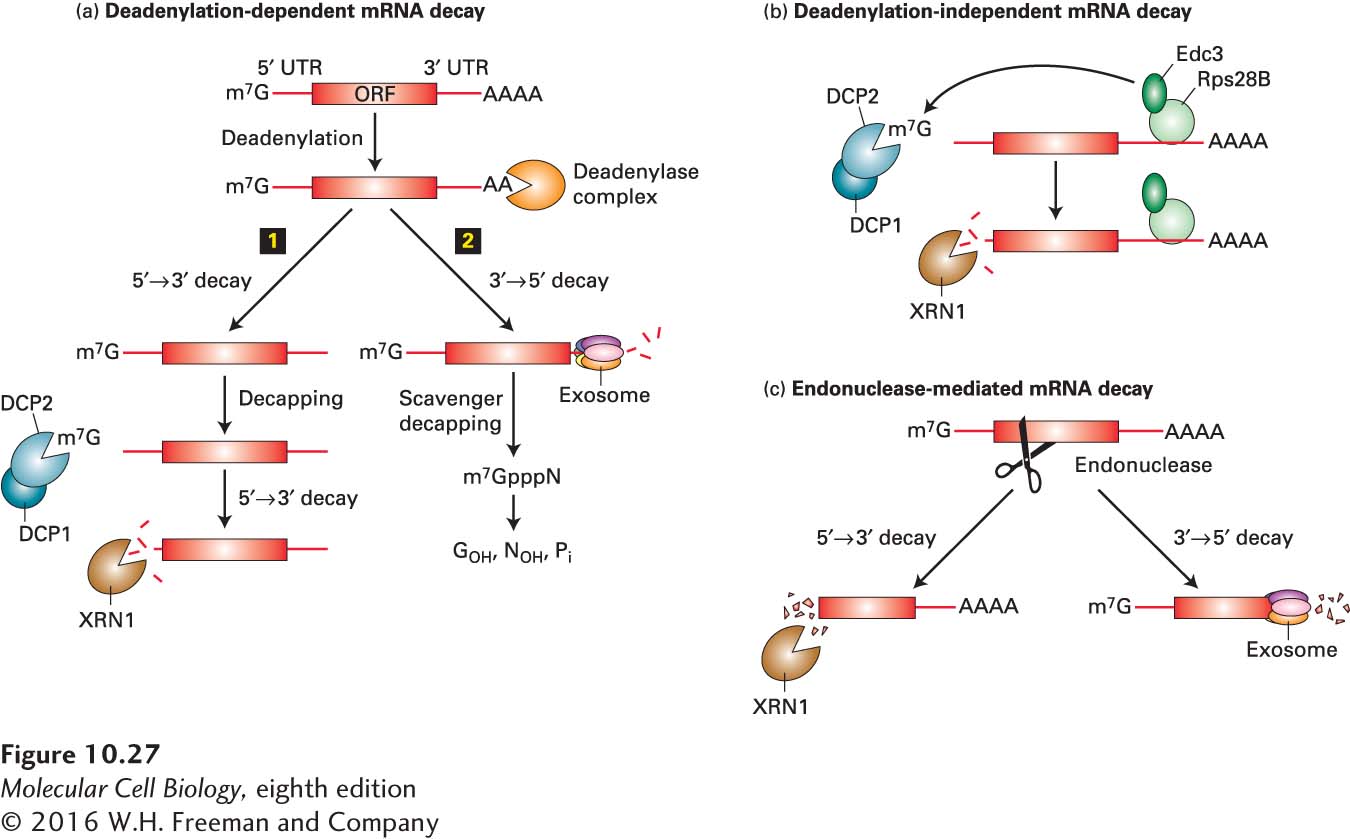
FIGURE 10- n- 5- A-