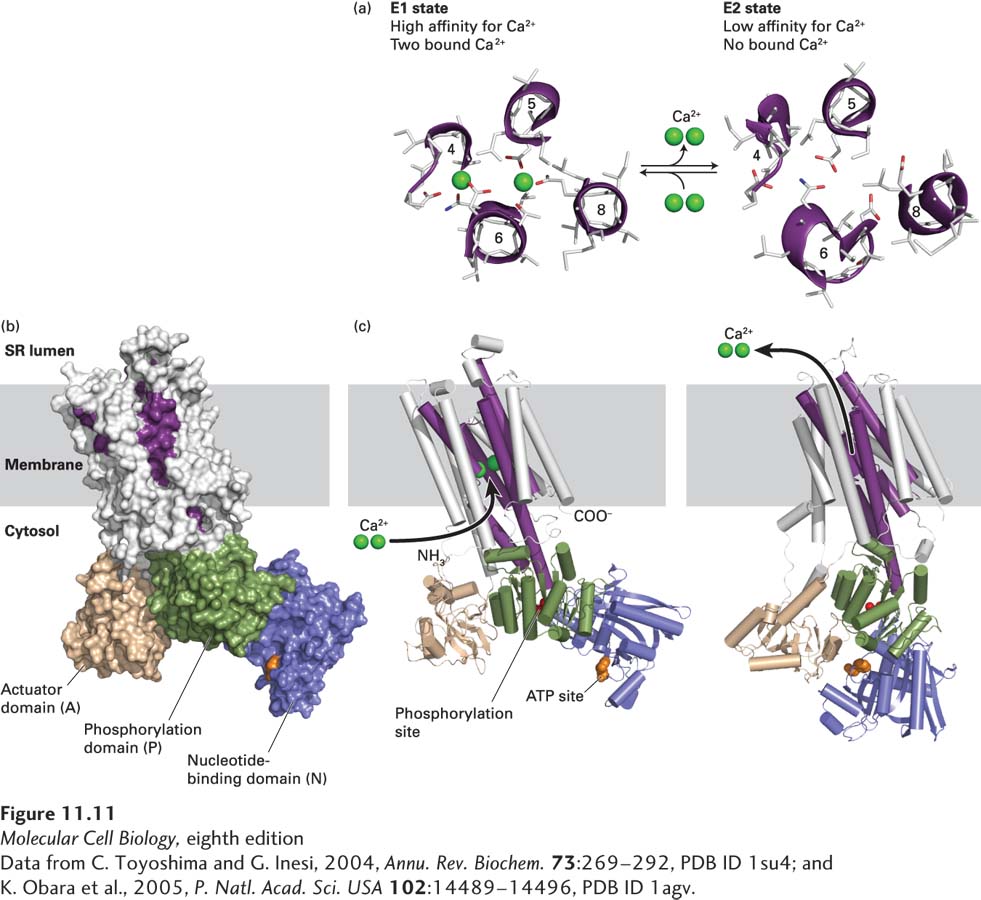
FIGURE 11- w- h- e- n- e- x- e- e- e-
[Data from C. Toyoshima and G. Inesi, 2004, Annu. Rev. Biochem. 73:269– 9–