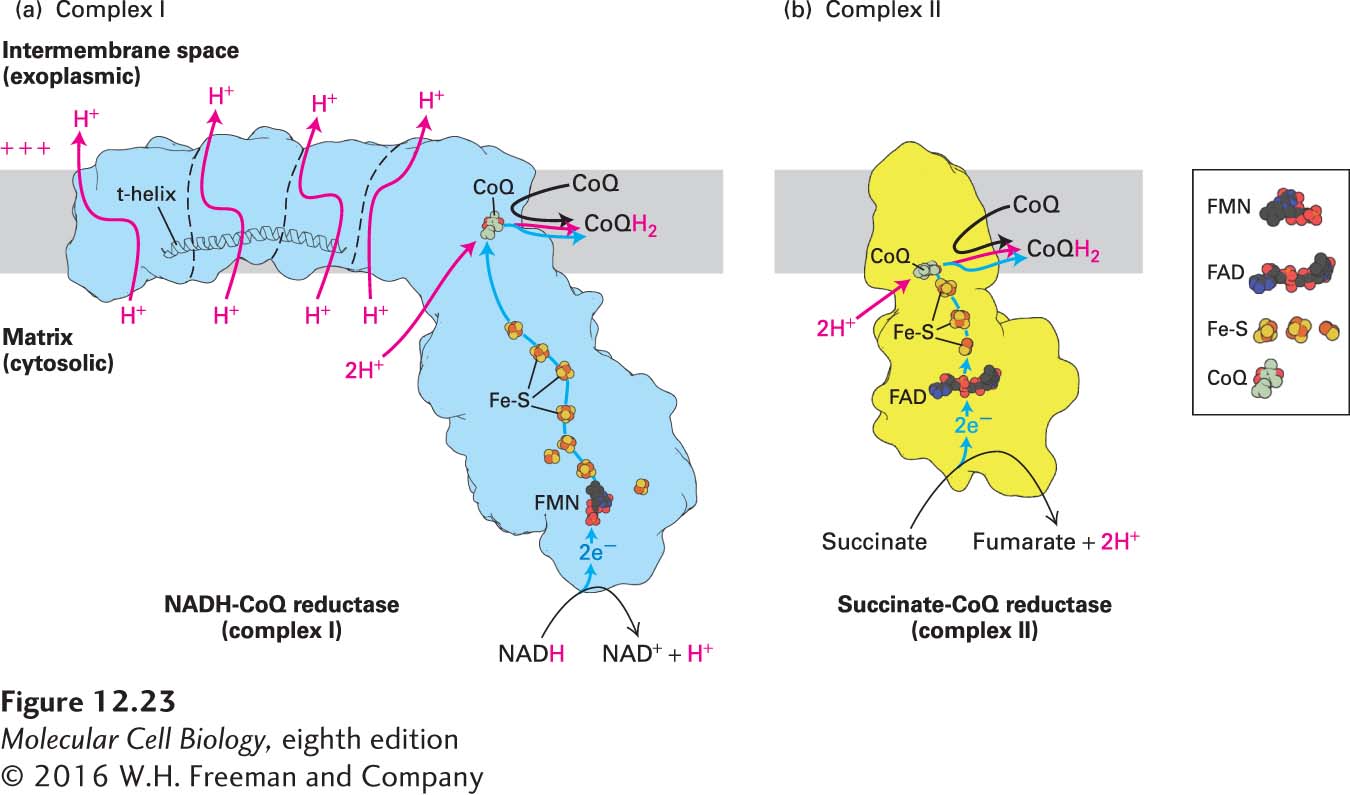
FIGURE 12- e- x- n- e- n- t- e- n- e-
[Part (a) data from V. Zickermann et al., 2015, Science 347:44– 3–