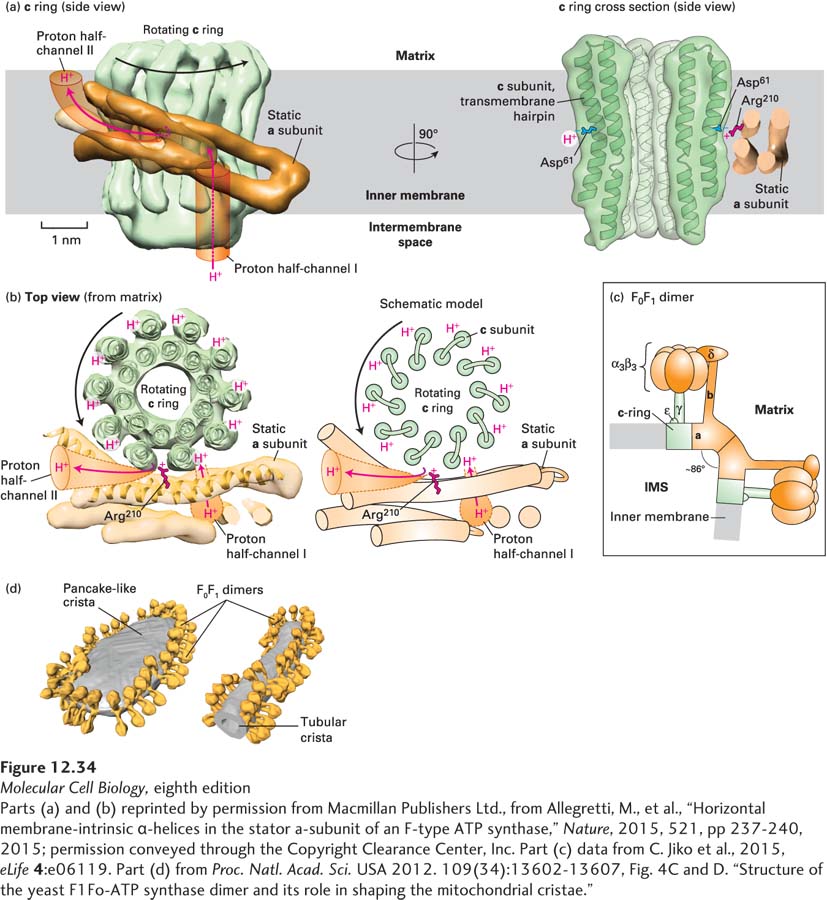
EXPERIMENTAL FIGURE 12- 34 High- resolution electron microscopy- based mechanism of proton translocation and bending of cristae membranes by ATP synthase. (a) and (b) The interface between the c ring (green) and a subunit (orange) of detergent- solubilized mitochondrial ATP synthase from the alga Polytomella sp., imaged by single- particle cryoelectron microscopy (~0.62 nm resolution), is shown (a) from within the plane of the inner mitochondrial membrane (side view) and (b) after a 90° rotation (top view). The movement of protons through half- channels I and II and the rotation of the c ring are described in detail in Figure 12- 31 . (a) Cross section through the c ring (right) shows that each c subunit is a transmembrane helical hairpin – two adjacent transmembrane α helices connected by a short nonhelical linker on the matrix side of the membrane. The negative side chain of the c subunit’s Asp61 in the middle of the membrane is thought to both serve as a binding site for translocating protons and interact with the side chain of the a subunit’s Arg210. (c) A model of the bovine heart mitochondrial ATP synthase is based on cryoelectron tomography and electron crystallographic image processing from crystalline ATP synthase in artificial membranes. Each F0F1 monomer bends the membrane by ~43° toward the intermembrane space (IMS), resulting in dimers bending the membrane by ~86°. The rotating c ring and γ and ε subinits are colored green, and the remaining static portions of the enzyme are shown in orange. (d) Cryoelectron tomographic image of frozen membranes from purified Saccharomyces cerevisiae (yeast) mitochondria. The surfaces of the ATP synthase complexes (orange) and the membrane (gray) show that the enzymes dimerize as in (c) and align into long rows that bend the membranes into characteristic tubular and flat, pancake- like cristae.
[Parts (a) and (b) reprinted by permission from Macmillan Publishers Ltd., from Allegretti, M., et al., “Horizontal membrane- intrinsic α-helices in the stator a- subunit of an F- type ATP synthase,” Nature, 2015, 521, pp 237- 240, 2015; permission conveyed through the Copyright Clearance Center, Inc. Part (c) data from C. Jiko et al., 2015, eLife 4:e06119. Part (d) from Proc. Natl. Acad. Sci. USA 2012. 109(34):13602- 13607, Fig. 4C and D. “Structure of the yeast F1Fo- ATP synthase dimer and its role in shaping the mitochondrial cristae.”]
[Leave] [Close]