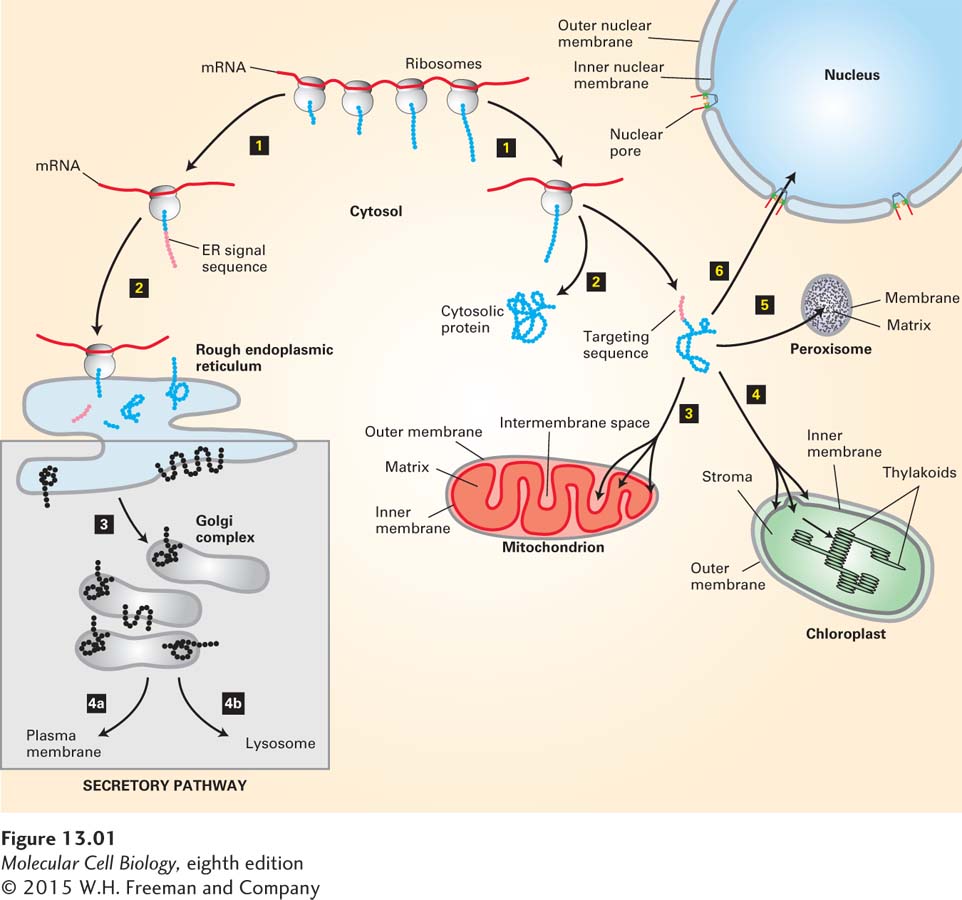
FIGURE 13- 1 Overview of major protein- sorting pathways in eukaryotes. All nuclear DNA– encoded mRNAs are translated on cytosolic ribosomes. Right (nonsecretory pathways): Synthesis of proteins lacking an ER signal sequence is completed on free ribosomes (step 1). Those proteins that contain no targeting sequence are released into the cytosol and remain there (step 2). Proteins with an organelle- specific targeting sequence (pink) are first released into the cytosol (step 2) but are then imported into mitochondria, chloroplasts, peroxisomes, or the nucleus (steps 3–6). Mitochondrial and chloroplast proteins typically pass through the outer and inner membranes to enter the matrix or stromal space, respectively. Other proteins are sorted to other subcompartments of these organelles by additional sorting steps. Nuclear proteins enter and exit through visible pores in the nuclear envelope. Left (secretory pathway): Ribosomes synthesizing nascent proteins in the secretory pathway are directed to the rough endoplasmic reticulum (ER) by an ER signal sequence (pink; steps 1 and 2). After translation is completed on the ER, these proteins can move via transport vesicles to the Golgi complex (step 3). Further sorting delivers proteins either to the plasma membrane or to lysosomes (step 4a or 4b). The vesicle- based processes underlying the secretory pathway (steps 3 and 4, shaded box) are discussed in Chapter 14.
[Leave] [Close]