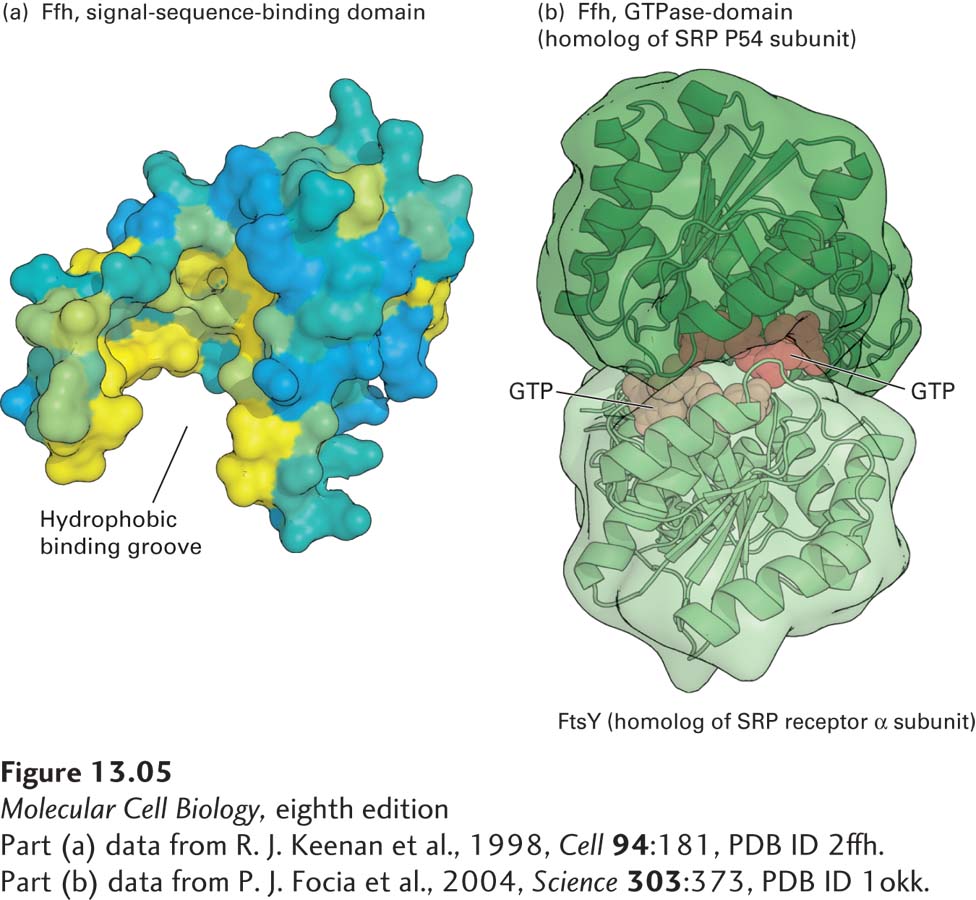
FIGURE 13- 5 Structure of the signal recognition particle (SRP). (a) The signal- sequence- binding domain: the bacterial Ffh protein is homologous to the portion of P54 that binds ER signal sequences in eukaryotes. This surface model shows the binding domain in Ffh, which contains a large cleft lined with hydrophobic amino acids (yellow) whose side chains interact with signal sequences. (b) GTP- and receptor- binding domain: the structure of GTP bound to FtsY (the archaeal homolog of the α subunit of the SRP receptor) and Ffh subunits from Thermus aquaticus illustrates how the interaction between these proteins is controlled by GTP binding and hydrolysis. Ffh and FtsY each can bind to one molecule of GTP, and when they bind to each other, the two bound molecules of GTP fit in the interface between the protein subunits and stabilize the dimer. Assembly of the pseudosymmetric dimer allows formation of two active sites for the hydrolysis of both bound GTP molecules. Hydrolysis to GDP destabilizes the interface, causing disassembly of the dimer.
[Part (a) data from R. J. Keenan et al., 1998, Cell 94:181, PDB ID 2ffh. Part (b) data from P. J. Focia et al., 2004, Science 303:373, PDB ID 1okk.]
[Leave] [Close]