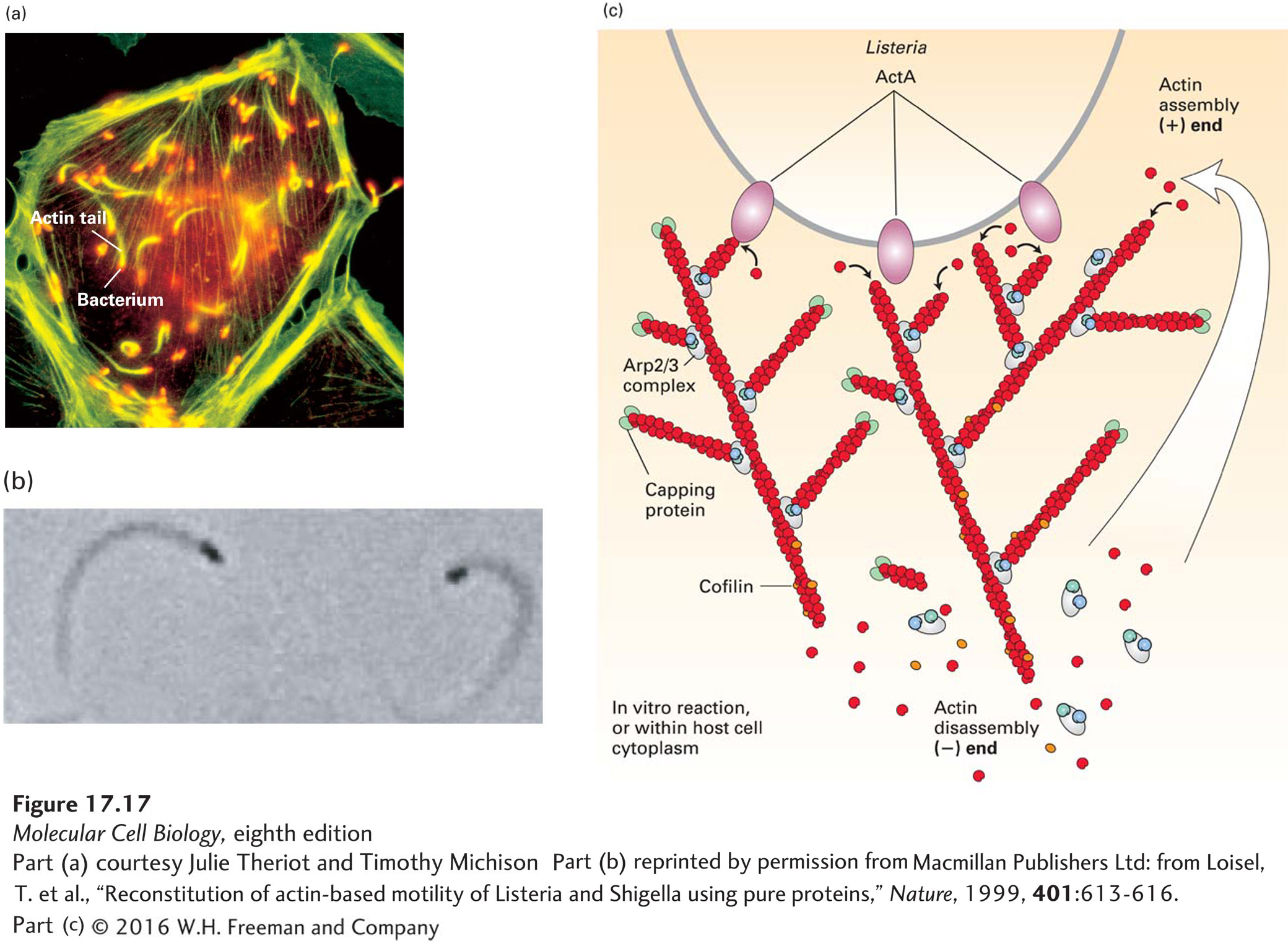
EXPERIMENTAL FIGURE 17- 17 Listeria uses the power of actin polymerization for intracellular movement. (a) Fluorescence microscopy of a cultured cell stained with an antibody to a bacterial surface protein (red) and fluorescent phalloidin to localize F- actin (green). Behind each Listeria bacterium is an actin “comet tail” that propels the bacterium forward by actin polymerization. When the bacterium runs into the plasma membrane, it pushes the membrane out into a structure like a filopodium, which protrudes into a neighboring cell. (b) Listeria motility can be reconstituted in vitro with bacteria and just four proteins: ATP– G- actin, Arp2/3 complex, CapZ, and cofilin. This phase- contrast micrograph shows bacteria (black), behind which are the phase- dense actin tails. (c) A model of how Listeria moves using just four proteins. The ActA protein on the bacterial cell surface activates the Arp2/3 complex to nucleate new filament assembly from preexisting filaments. Filaments grow at their (+) end until capped by CapZ. Actin is recycled through the action of cofilin, which enhances depolymerization at the (−) end of the filaments. In this way, polymerization is confined to the back of the bacterium and propels it forward. Although not essential for motility in vitro, the protein VASP (not shown) binds ActA in vivo to enhance motility, as described in the text.
[Part (a) courtesy Julie Theriot and Timothy Michison; part (b) reprinted by permission from Macmillan Publishers Ltd: from Loisel, T. et al., “Reconstitution of actin- based motility of Listeria and Shigella using pure proteins,” Nature, 1999, 401:613- 616.]
[Leave] [Close]