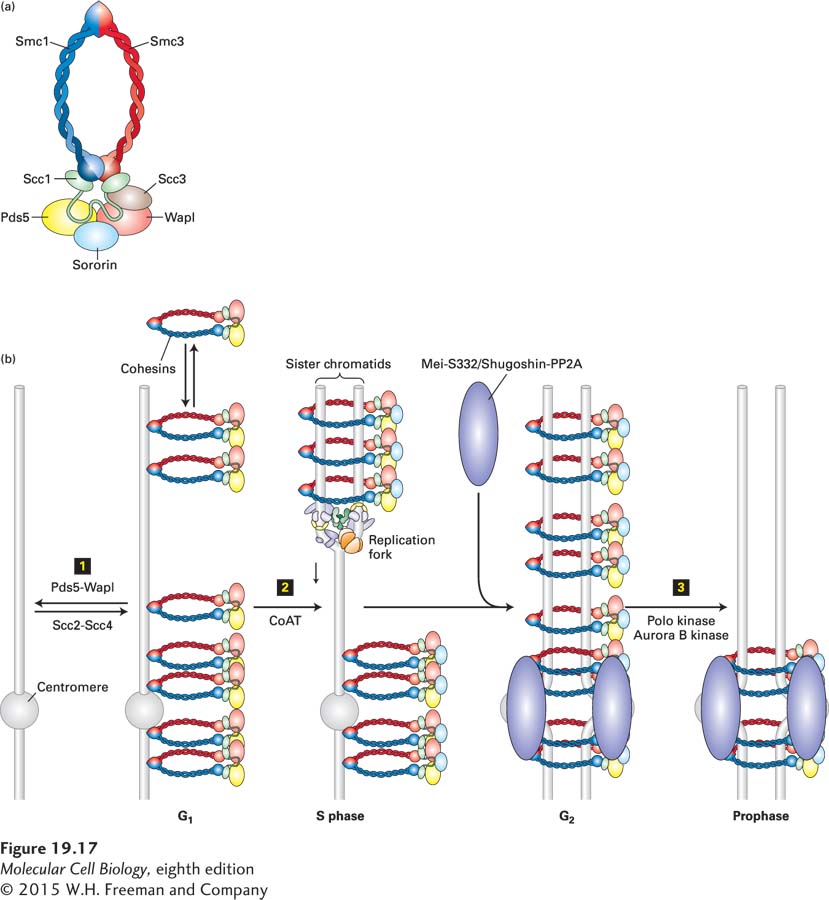
FIGURE 19- n- 2- 5- i- 5- i-