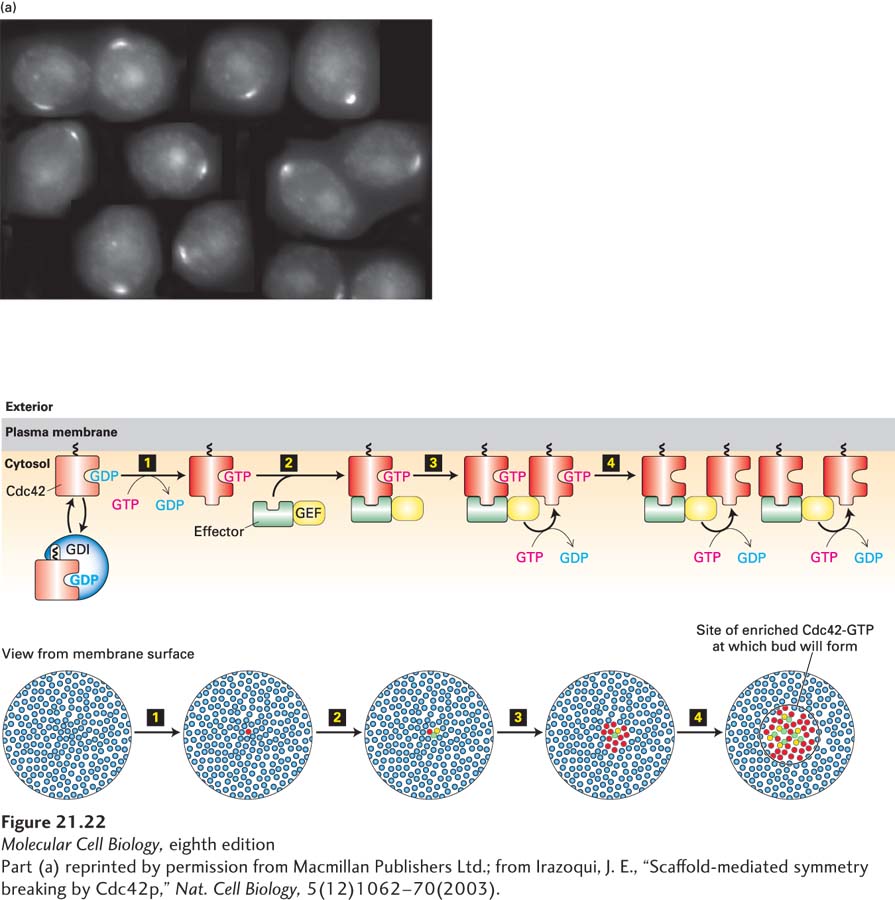
FIGURE 21- 22 The intrinsic polarity program of budding yeast involves a positive feedback loop for activation of the GTPase Cdc42. (a) Diploid yeast lacking polarity cues show polarized Cdc42, visualized here by immunofluorescence microscopy, when they are about to assemble a bud. The cells were treated with drugs to disassemble both actin filaments and microtubules to show that polarization of Cdc42 is not dependent on these cytoskeletal filaments. (b) Positive feedback loop for activation of Cdc42. Inactive Cdc42·GDP is in equilibrium between a cytosolic pool of complexes with the guanine nucleotide dissociation inhibitor (GDI) and a membrane- bound pool. Step 1: One of the membrane- associated Cdc42·GDP proteins may spontaneously become an activated Cdc42·GTP. Step 2: Active Cdc42·GTP recruits a complex containing the guanine nucleotide exchange factor (GEF). Step 3: The recruited GEF now locally converts more Cdc42·GDP to Cdc42·GTP. Step 4: This active Cdc42·GTP recruits more GEF, thus driving a positive feedback loop that results in the local accumulation of Cdc42·GTP. See C.-F. Wu and D. Lew, 2013, Trends Cell Biol. 23:476.
[Part (a) reprinted by permission from Macmillan Publishers Ltd.; from Irazoqui, J. E., “Scaffold- mediated symmetry breaking by Cdc42p,” Nat. Cell Biology, 5(12)1062– 70(2003).]
[Leave] [Close]