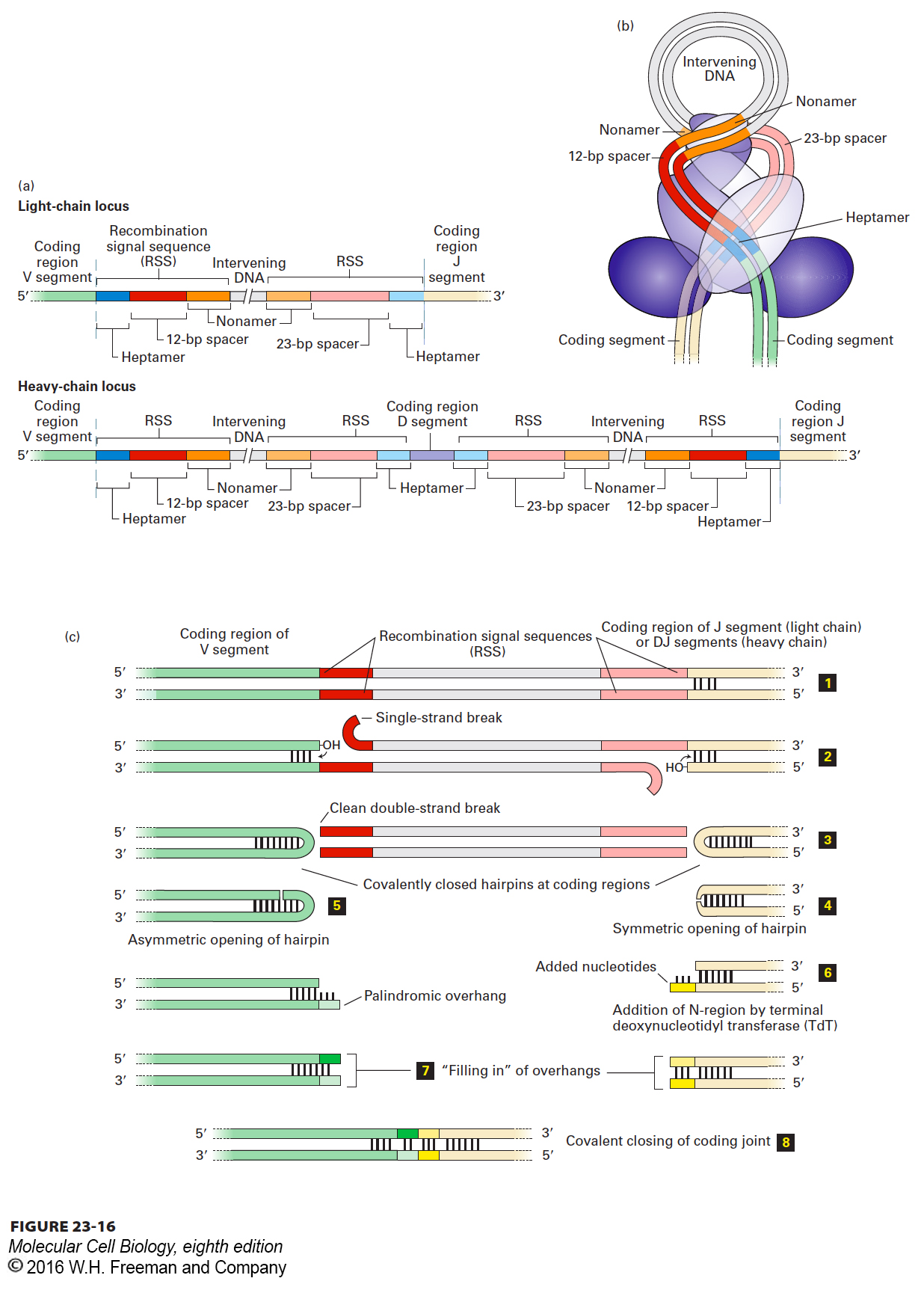
FIGURE 23- t- y- y- t- 2- 3- m- e- e- y- e- N- N-